Abstract
Background
Biting midges of the genus Culicoides (Diptera: Ceratopogonidae) transmit pathogens that cause important diseases. No effective technique has been found to properly control either Culicoides spp. abundance or their likelihood to transmit pathogens. Endosymbionts, particularly Wolbachia, represent powerful alternatives to control arthropods of health interest. In arthropods, Wolbachia can reduce vector fitness and vector’s pathogen transmission capacity, thus being a potential target for population reduction and replacement strategies.
Results
The presence of Wolbachia and Cardinium endosymbionts was screened in Spanish Culicoides spp. populations at livestock premises and natural habitats. The first detection of Wolbachia-infected Culicoides spp. in Europe is reported. The putative Palaearctic vectors for bluetongue and Schmallenberg diseases, C. imicola, C. obsoletus (s.s.) and C. pulicaris (s.l.), were infected with Wolbachia. Four genetic clusters of closely-related Wolbachia strains from A and B supergroups were detected infecting Culicoides. Cardinium strain of the C-group was detected in C. obsoletus (s.l.). Both endosymbionts, Wolbachia and Cardinium, were detected in Culicoides species of minor epidemiological relevance as well. Higher prevalence of Wolbachia infection was detected in natural habitats, while livestock premises lead to higher prevalence of Cardinium. Significant differences in the prevalence of Wolbachia, but not Cardinium, were also detected between some Culicoides species and between locations.
Conclusions
The presence of Wolbachia and Cardinium endosymbionts in Culicoides is expected to trigger new research towards the control of Culicoides-transmitted diseases. The results of the present study could have an impact beyond the Culicoides arena because successful Wolbachia transfection is possible even across genus and species barriers.
Similar content being viewed by others
Background
Biting midges of the genus Culicoides (Diptera: Ceratopogonidae) are well known worldwide for transmitting pathogens that cause important diseases. In Europe, Culicoides became a major concern for spreading the largest Bluetongue (BT) epizootics ever recorded [1]. Since 1998, BT was reintroduced, and recurrent outbreaks are still ongoing. Through BT disease, Culicoides provoked major economic losses with important disruption of international animal trade [2, 3]. In addition, Culicoides are involved in the spread of other important arboviral diseases such as African horse sickness [4], Epizootic haemorrhagic disease [4], and the recently emerged Schmallenberg disease [5, 6]. Culicoides are also vectors for pathogens of different aetiology, such as filarial worms and protozoa [4]. To date, no effective vector control technique or approach to limit the likelihood of pathogen transmission has been found for Culicoides [7, 8].
Endosymbiotic bacteria are naturally found in insects [9]. In recent years, the growing number of field screenings in several arthropod phyla led to an increase in the reported endosymbiont prevalence in arthropods [10]. A meta-analysis performed by Hilgenboecker et al. [11] suggested up to 66% of all insect species were infected with Wolbachia (class Alphaproteobacteria, order Rickettsiales). Wolbachia is an obligate intracellular endosymbiotic bacteria present in a wide range of arthropods and filarial nematodes worldwide. Wolbachia and other bacterial endosymbionts are well known as master manipulators of arthropod host reproduction [12]. Such endosymbionts can manipulate host reproduction by inducing cytoplasmic incompatibility of host gametes, feminization of genetic males, parthenogenesis and male-killing [13, 12]. The endosymbiont Cardinium (Cytophaga-Flavobacterium-Bacteroides) is another well-characterised endosymbiont of arthropods. Cardinium has a lower infection prevalence and is restricted, apparently, to fewer taxonomic groups [14].
Research on arthropod endosymbionts is strongly consolidated for mosquitoes [15]. This challenge has been addressed for other important arthropod vectors such as tsetse flies [16]. Despite its potential interest, the prevalence of endosymbionts across Culicoides communities and ecosystems has been scarcely studied. However, recent studies have endorsed the presence of endosymbionts in Culicoides. First attempts were performed by Nakamura et al. [17] in Asia. The authors reported, for the first time, Culicoides midges infected with Wolbachia and “Candidatus Cardinium hertigii” (Bacteroidetes). The latter led to the description of a new Cardinium group (group C). Soon after, Morag et al. [18] described Cardinium-infected Culicoides in Israel. More recently, Lewis et al. [19] detected the same endosymbiont in Culicoides in the United Kingdom. Cardinium DNA sequences of the strains reported in the three previous studies are highly conserved. More recently, Wolbachia and Cardinium were detected in Culicoides species from the Australasian region and Africa [20]. A recent study confirmed the absence of Wolbachia in feminized males of the species C. circumscriptus collected at ten populations from Spain [21]. To date, endosymbionts of the genus Wolbachia remain undetected in European Culicoides communities.
Use of endosymbionts, Wolbachia particularly, is a promising approach for controlling the dynamics of some arthropod-transmitted pathogens under certain scenarios. They have the potential to modulate major parameters of arthropod vectorial capacity. Thus, infection with certain Wolbachia strains can lead to a decrease in arthropod survival as shown in Aedes aegypti [22]. Moreover, Wolbachia can influence arthropod’s vector competence of important pathogens. For example, Wolbachia-infected Anopheles stephensi became refractory to infection with Plasmodium falciparum [23]. Wolbachia has been studied as well for its potential to introduce transgenes into arthropod natural populations [24], and more interestingly within vectors of arthropod borne diseases [25]. Wolbachia and (indirectly) the mitochondrial genome of its host can rapidly invade and establish through uninfected populations by manipulating its host-reproduction with the mechanisms mentioned above [13].
The present study examined whether Wolbachia and Cardinium bacterial endosymbionts naturally infected Culicoides species communities across Spanish ecosystems. Once confirmed, Wolbachia and Cardinium strains were genotyped. Then, the effect of Culicoides species, geographical origin and habitat type on endosymbiont prevalence was assessed.
Methods
Sampling
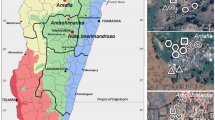
Biting midges were captured at four Spanish NUTS2 (Nomenclature des unités territoriales statistiques) administrative units: Catalonia, Asturias, Castilla La Mancha and Andalucía. Twenty-four sites were sampled across the Iberian Peninsula (Fig. 1, Table 1). At each NUTS2, sampling sites included two habitat types, livestock premises and natural habitats (Table 1). Natural habitats consisted of forestry areas usually located more than 1 km far from the closest livestock farm. Culicoides collections were made between the years 2009–2012. Biting midges were trapped using Center for Disease Control (CDC) Miniature ultraviolet (UV)-light traps (model 912, John W. Hock Company, Gainesville, USA) in soapy water during the night. Culicoides were recovered the day after and transferred to 70% ethanol. Culicoides were identified under a stereomicroscope (Nikon model SMZ) according to their pattern of wing pigmentation [26] and stored frozen (-20 °C) for further downstream processing. A map including Culicoides collection sites and endosymbionts spatial distribution was prepared with Quantum GIS software [27] using the coordinate reference System (CRS) EPSG:4326, WGS 84.
Sampling sites in Spain where Culicoides spp. were tested for the presence of endosymbionts. Sampling sites: 1, Almoraima; 2, Amposta; 3, Aramunt; 4, Bonastre; 5, Brunyola; 6, Caldes de Malavella; 7, Canyamars; 8, Colunga; 9, Garcia; 10, Juanar; 11, La Galera; 12, La Morera; 13, Massanas; 14, Piera; 15, Pozo Estanco; 16, Proaza; 17, Quintos de Mora; 18, Roda de Barà; 19, Sant Iscle de Vallalta; 20, Sant Just Desvern; 21, Santa Clara; 22, Susqueda; 23, Terrades; 24, Vilanova de la Muga
DNA extraction, PCR and sequencing
Crude homogenates of individual female midges were prepared in 200 μl phosphate buffered saline (PBS), using glass beads in a Fastprep® (MP Biomedicals, Solon, USA) at a speed of 5 m/s. A fraction of 10 homogenates (20 μl each), of the same species and collection site, were pooled for DNA extraction. DNA extractions, both from pools or individuals, were performed using a commercial kit (DNeasy Blood and Tissue Kit, Qiagen, Crawley, UK) following the manufacturer’s instructions, with a final elution volume of 100 μl. Two positive controls were used. The first was obtained from a Culicoides infected with Wolbachia. The second was obtained from a Culicoides infected with Cardinium. A non-infected Culicoides was used as negative control.
Presence of endosymbiotic bacteria of the genus Wolbachia and Cardinium was assessed using specific single polymerase chain reaction (PCR). Briefly, the presence of Wolbachia was tested by amplification of a fragment of the wsp (Wolbachia surface protein) gene delimited by primers wsp81F and wsp691R [28]. The presence of “Candidatus Cardinium heretgii” was tested by amplification of a fragment of the 16S rDNA delimited by primers CLO-f1 and CLO-r1 [26]. The first screening was performed in pools of 10 Culicoides. When a pool was confirmed positive, the initial crude homogenates, corresponding to individual Culicoides in the positive pooled sample were used for individual DNA extraction and endosymbiont PCR detection. Positive individuals from the Obsoletus group were identified to species using a cytochrome oxidase subunit I (cox1) gene specific PCR test [29, 30]. PCR reactions were carried out using the GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). Negative controls were included in every PCR. PCR products were confirmed by ethidium bromide staining after electrophoresis on 2% (w/v) agarose 1× TAE gel run at 10 V/cm for 1 h.
When considered necessary, positive samples were sequenced. PCR products were purified, and DNA purified products were sequenced on both strands using Big Dye Terminator version 3.1 cycle sequencing kit (Applied Biosystems) and analysed on an ABI PRISM 3730 Automated sequencer (Applied Biosystems).
Phylogenetic analysis
DNA sequences were edited using Bioedit sequence alignment editor software (version 5.0.9. for Windows [31]) and aligned with ClustalW Multiple alignment option without manual optimization. Phylogenetic and molecular evolutionary analyses were conducted using MEGA6 software [32]. Phylogenetic analysis was inferred using Maximum Likelihood (ML) method incorporating best-fit models of sequence evolution determined using the Akaike information criterion with a resampling nodal support of 1000 bootstrap replicates. Best-fit models were Tamura 3-parameter (T92 + G) for Wolbachia dataset, and Kimura 2-parameter Gamma corrected (K2 + G) for Cardinium. Sequences published at the National Center for Biotechnology Information (NCBI) were used to better determine the evolutionary relationship of Wolbachia and Cardinium isolates (Additional file 1: Table S1).
Statistical analyses
The prevalence of Wolbachia and Cardinium infection was calculated for Culicoides species, site, geographical region and habitat. Differences in infection prevalence were tested using the two tailed Fisher’s exact test [18]. Differences among species were tested separately in sites where more than one Culicoides species was captured, and at least in one of the species endosymbiont infection was detected. Differences among sites of the same administrative unit and habitat type were tested separately in species of Culicoides that were trapped in more than one site and at least in one of these sites positive infections were detected. Differences between habitats were tested within geographical regions both separating by Culicoides species and grouping all species together, and considering the whole sample together as well. All these analyses were performed separately for the prevalence of Wolbachia and Cardinium infections.
Results
PCR screening for Wolbachia and Cardinium allowed the detection of endosymbionts in several Culicoides populations (Fig. 1). The most epidemiologically relevant Culicoides species in Europe were infected with Wolbachia and Cardinium (Table 2). No double infection was detected in the 1050 Culicoides analysed. However, Wolbachia and Cardinium were found in sympatry in three out of the 24 sampled sites (Fig. 1).
Wolbachia screening
Wolbachia infection was present in some of the putative vectors of bluetongue (BTV) and Schmallenberg (SBV) viruses in Europe: C. imicola, C. obsoletus (s.l.) and C. pulicaris (s.l.) Wolbachia infection was also detected in C. vexans, C. kibunensis and C. heteroclitus.
The prevalence of Wolbachia infection detected at metapopulation scale was low for the species with a representative geographical screening at the population level: C. imicola, C. obsoletus (s.l.) and C. pulicaris (s.l.). In C. imicola, 6 out of 256 individuals were positive with a prevalence ratio (PR) of 0.023. In C. obsoletus (s.l.), 2 of 466 individuals tested were positive (PR = 0.004). Conversely, infection prevalence in C. pulicaris (s.l.) was higher, with 56 positive individuals out of the 309 tested (PR = 0.18). These results were supported by statistical tests, which indicated that prevalence of infection differed between some Culicoides species. The prevalence of Wolbachia in C. pulicaris (s.l.) was significantly higher than in C. imicola and C. obsoletus in two out of the three sites tested (Almoraima: P < 0.001 in both specific comparisons; Quintos de Mora: P < 0.05 in both specific comparisons). However, the prevalence of Wolbachia did not differ between C. obsoletus and C. imicola in any of the three sites where both species were detected in sympatry.
Our screening indicated that Wolbachia infection had a heterogeneous spatial distribution (Fig. 1, Table 2, Additional file 2: Table S2). The prevalence of Wolbachia infection in particular Culicoides species even differed among sites of the same geographical region. Two out of three populations of C. imicola from Andalucía, Almoraima and Santa Clara, were infected with Wolbachia. Although the prevalence of Wolbachia infection did not significantly differ among the three populations from Andalucía, differences between Almoraima (PR = 0.10) and Juanar (PR = 0.00) were almost significant (P = 0.058). None of the C. imicola populations screened in Castilla La Mancha, or Catalonia tested positive. Culicoides imicola was not collected in Asturias. Wolbachia infection in C. obsoletus (s.l.) was evident in one out of three populations of Asturias (Proaza). However, infection prevalence did not differ significantly among them. The two Culicoides females infected with Wolbachia in Proaza were genetically identified as C. obsoletus (s.s.). None of the C. obsoletus (s.l.) populations from Andalucía, Castilla la Mancha or Catalonia, tested positive. Three populations of C. pulicaris (s.l.) from Andalucía (Almoraima), Castilla La Mancha (Quintos de Mora), and Asturias (Proaza) were infected with Wolbachia. The population of C. pulicaris (s.l.) from Almoraima showed the highest infection prevalence detected in the study, with more than 90% of specimens positive for Wolbachia. The prevalence of Wolbachia infection detected in Almoraima was significantly higher than in the remaining populations of C. pulicaris (s.l.), including another population from Andalucía, Juanar (P < 0.001), in which Wolbachia was not detected.
An effect of habitat type was also detected, with a higher prevalence of Wolbachia infection in natural habitats than in livestock premises. However, the effect of habitat could not be detected in individual species. When grouping all the Culicoides species the prevalence of Wolbachia infection was statistically higher in natural habitats compared to that found in livestock premises in Andalucía (P < 0.001) and when all regions were considered simultaneously (P < 0.001).
Cardinium screening
Cardinium infection was present in a wide range of species: C. obsoletus (s.l.), C. festivipennis, C. flavipulicaris, C. haranti, C. maritimus, C. minutissimus, C. newsteadi, C. punctatus and C. sahariensis.
Conversely, to what was observed for Wolbachia infections, the prevalence of Cardinium infection differed neither between Culicoides species nor between sites, mainly because of the scarce number of infected specimens. Prevalence of Cardinium was low for the C. obsoletus (s.l.) metapopulation analyzed (PR = 0.004; 454 females tested). The endosymbiont was found in two C. obsoletus (s.l.) populations. The first population, Colunga, was located in a forestry area of Asturias (PR = 0.02; 51 females tested). The second, Massanes, was present at a livestock farm in Catalonia (PR = 0.01; 70 females tested). Prevalence of Cardinium infection in other Culicoides species might be biased because there was a low sample size (Additional file 2: Table S2). As observed for Wolbachia, a significant effect of habitat type was detected for the prevalence of Cardinium infection. In this case, however, infection prevalence was higher in livestock premises than in natural habitats. Thus, when grouping all the Culicoides species, the prevalence of infection was statistically higher in livestock premises than in natural habitats in Catalonia (P < 0.01) and when all regions were considered simultaneously (P < 0.001).
Phylogenetic analyses of endosymbionts
Wolbachia
Wolbachia DNA partial sequences of the wsp gene were obtained for 23 Wolbachia-infected Culicoides out of 67 (GenBank: MF179641–MF179663). The DNA dataset was representative of all Culicoides taxonomic groups that were found infected with Wolbachia in each geographical area (Fig. 1, Table 2). Nine haplotypes were detected among the 23 wsp gene sequences, with haplotype diversity (Hd) of 0.787 and nucleotide diversity (π) of 0.0916. Phylogenetic analyses placed wsp sequences derived from Spanish Culicoides within the Wolbachia supergroups A (n = 4) and B (n = 19). Spanish isolates were grouped into four clades, two into supergroup A and two into supergroup B (Fig. 2). Wolbachia supergroup A sequences were grouped in two clades, III-WA and IV-WA, and were from Culicoides collected in Andalucía (Table 3). Isolates of both clades infected C. imicola whereas a single isolate of IV-WA infected C. pulicaris (s.l.) as well. Wolbachia supergroup B sequences were grouped in two distant clades, I-WB and II-WB (Fig. 2). The first clade (I-WB) had Wolbachia sequences isolated from C. imicola, C. kibunensis, C. pulicaris (s.l.) and C. vexans collected at Andalucía, Catalonia and Castilla la Mancha (Table 3). Clade II-WB was representative of C. pulicaris (s.l.) and C. obsoletus (s.l.) infections (from Asturias and Castilla la Mancha) and contained sequences previously detected in Australian Culicoides [20].
Cladogram for Wolbachia phylogenetic relationships inferred from wsp gene sequences; sequences already known from Culicoides are indicated with white diamonds; new wsp gene sequences of Wolbachia-infected Culicoides obtained in the present study are indicated with black diamonds. Accession numbers followed by host species are shown at terminal nodes. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Maximum likelihood (ML) bootstrap support values over 50% are displayed on the nodes. Abbreviations: I-WB, clade I-Wolbachia supergroup B; II-WB, clade II-Wolbachia supergroup B; III-WA, clade III-Wolbachia supergroup A; IV-WA, clade IV-Wolbachia supergroup A; And, Andalucía; Ast, Asturias; Cas, Castilla la Mancha; Cat, Catalonia
Sympatric infections with Wolbachia from clades III-WA, IV-WA and I-WB were detected at a C. imicola population from Almoraima (Andalucía; Table 3). At the same collection site, the C. pulicaris (s.l.) population was infected with Wolbachia from clades IV-WA and I-WB. Moreover, C. pulicaris (s.l.) from Quintos de Mora (Castilla la Mancha) were infected with Wolbachia belonging to clades I-WB and II-WB.
Wolbachia sequences previously detected in Australian Culicoides spp. [20] were segregated into two separated clades within supergroup B and a third clade into supergroup A (Fig. 2). The first clade contained three sequences of the species C. narrabeensis (GenBank: KR026947–KR026949) and was the phylogenetically related to clade I-WB (evolutionary distance, ED = 0.161). A sequence from C. narrabeensis (GenBank: KR026950) and one from C. brevitarsis (GenBank: KR026943) were included into clade II-WB. The last group of Wolbachia wsp gene sequences detected in Australian Culicoides were placed in a third clade into supergroup A (C. brevitarsis, GenBank: KR026944, KR026945; C. antennalis, KR026942; C. bundyensis, KR026946; C. parvimaculatus, KR026951). This clade was relatively distant from clades III-WA (ED = 0.182) and IV-WA (ED = 0.206).
Within each of the four clades, nucleotide variation was minor or absent regardless Culicoides species or geographical location (Fig. 2). Within clades, evolutionary distances were 0.001 (I-WB), 0.004 (II-WB) and 0 (IV-WA).
Cardinium
A fragment (416 bp) of the 16S ribosomal DNA gene was obtained from 8 Cardinium-infected Culicoides midges out of 12 (GenBank: MF188893–MF188900). The DNA dataset was representative of all Culicoides taxonomic groups that were infected with Cardinium in each geographical area (Fig. 1, Table 2). Nucleotide diversity among the eight Cardinium 16S RNA gene sequences was low (π = 0.0008), with two haplotypes differing by a singleton (Hd = 0.333). When analysed with previously published sequences of Cardinium strains detected in Culicoides (n = 26, Additional file 1: Table S1), the new sequence data exhibited low genetic variation (π = 0.0073, Hd = 0.504). However, the cladogram showed that all Cardinium sequences derived from Culicoides biting midges (n = 32) were grouped in a single clade (Fig. 3). This clade formed the recently described Cardinium C-group, attributed exclusively to Culicoides infections [17]. Additional Cardinium sequences from other arthropod groups and nematodes were used to infer better phylogenetic relationships among Cardinium. Cardinium B-group was derived from a strain detected in nematodes. Cardinium A-group was constituted by Cardinium strains isolated from different arthropod groups (Fig. 3).
Cladogram for Cardinium phylogenetic relationships inferred from 16S sequences. Cardinium 16S rDNA sequences obtained in the present study are indicated with black diamonds. Accession numbers followed by host species are shown at terminal nodes. A. asiaticus has been used as outgroup to root the cladogram. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Maximum likelihood (ML) bootstrap support values over 50% are displayed on the nodes
Discussion
To the best of our knowledge, the present study represents the first detection of Wolbachia infecting Culicoides in Europe. Endosymbionts of the genera Wolbachia and Cardinium have been detected in the putative Palaearctic vectors of BTV and SBV.
Previous studies have detected Cardinium endosymbionts in relevant Palaearctic Culicoides species, i.e. C. imicola [18], C. pulicaris and C. punctatus [19], usually with an intermediate prevalence. The role for Cardinium infection in arthropod’s fitness remains uncertain to date. In Culicoides, no difference in survival rate was attributed to Cardinium infection for C. imicola in laboratory conditions [33]. However, the present study widens the known Cardinium infection host range within the genus Culicoides. The most relevant species infected with Cardinium was C. obsoletus (s.l.). The absence of Cardinium infection in C. imicola contrasts with results obtained in Israel [17] and Africa [19]. However, Cardinium infection might occur in C. imicola non-tested populations.
Wolbachia was present in Culicoides species of major epidemiological relevance: C. imicola, C. obsoletus (s.l.) and C. pulicaris (s.l.). However, Wolbachia was detected in other Culicoides species of less epidemiological relevance as well. Statistical analyses showed that the prevalence of Wolbachia infection diverges among some species of Culicoides. In all cases, the species that showed the highest infection prevalence was C. pulicaris (s.l.), with differences in prevalence being statistically significant when compared both with C. imicola and C. obsoletus (s.l.). While the infection prevalence of these two species did not diverge when compared at the same site, differences were marginally significant in the Andalucía region (result not shown). Prevalence of Wolbachia infection did not only diverge among species, but also among localities. The species C. pulicaris (s.l.) showed the largest geographical range for Wolbachia infection. Wolbachia-infection in C. pulicaris (s.l.) was present in Andalucía, Asturias and Castilla La Mancha. Also, one of the populations (C. pulicaris from Almoraima; Additional file 2: Table S2) exhibited a high prevalence of infection, being close to fixation. Statistical analyses also indicated significant differences between habitats. In particular, prevalence of Wolbachia infection was higher in natural habitats than in livestock premises. However, these differences were mainly due to the high prevalence detected in C. pulicaris (s.l.) from one of the natural habitats (Almoraima). In contrast, the prevalence of Cardinium infection was higher in livestock premises than in natural habitats. These results might suggest a negative association between the two endosymbionts in the Culicoides communities. In fact, no double infection was detected among Culicoides, and although sympatric Wolbachia and Cardinium infections were detected, none of the infections affected the same Culicoides species. Further analyses are needed to ensure a possible effect of habitat type on the prevalence of endosymbionts and their relationship.
The large geographical range for Wolbachia-infected Culicoides detected in Spain suggests that infections may be found in other regions of Europe. A similar situation would be expected for Cardinium infections. Also, the recently discovered presence of Wolbachia or Cardinium low level infections in Culicoides [20] strongly suggests the possibility for a higher prevalence of infection than the one here reported. The presence of such low level infections in Culicoides, even beyond the diagnostic sensitivity of the technique we used, cannot be ruled out. Future studies should use more sensitive techniques to detect low-level infections to ascertain Wolbachia (and other endosymbionts) incidence and prevalence across Culicoides populations.
The genus Wolbachia is highly diverse. Most insect-infecting Wolbachia belong to supergroups A and B, while C and D are found in filarial nematodes, and indirectly in arthropods harbouring such nematodes [13]. Some Wolbachia isolates we detected represented new strains detected in Culicoides. wsp gene sequences were grouped in four clades within Wolbachia supergroups A and B. Based on the cladogram, the diversity of Wolbachia infecting Culicoides is expected to be complex at a global scale. The phylogenetic analysis including sequences for the few known Wolbachia infecting Culicoides revealed up to six well-defined clades. Some of the sequences were highly divergent as shown by the separation of the clades into different Wolbachia supergroups (A or B). Phylogenetic analyses based on 16S rDNA showed that Cardinium strains detected in Culicoides were almost identical, as was reported in previous studies [17,18,19]. The isolates detected in Spain consolidate the presence of the new Cardinium C-group that apparently is specific of Culicoides biting midges at a global scale.
Endosymbiont based research, especially involving Wolbachia, has proven to be a promising technique to control some arthropod borne diseases. One of the most successful research lines involves the use of the life-shortening wMelPop Wolbachia strain. This strain has shown an important reduction of the lifespan of certain arthropods [34]. Moreover, the wMel Wolbachia strain has proven to induce refractoriness towards certain pathogens in mosquitoes [35, 36]. Thus, Wolbachia strains of Culicoides need to be functionally screened for properties targeting the reduction of either vector’s fitness or pathogen transmission. Wolbachia horizontal transmission between different host species has been proposed because of the phylogenetic incongruence between hosts and Wolbachia strains [37]. This was proven to be feasible by inducing stable infections in naïve arthropod populations [38]. Therefore, new findings could have an important impact beyond Culicoides arena because of the successful transfection of Wolbachia even across genus and species barriers.
Conclusions
The presence of natural infections of Wolbachia and Cardinium endosymbionts in Culicoides deserves attention. The finding might represent the starting point to address new research for the control of Culicoides-transmitted diseases. Wolbachia can control vector fitness and vector’s pathogen transmission, thus being a potential target for population reduction and replacement strategies. The results of the present study could have an impact beyond Culicoides arena because successful Wolbachia transfection is possible across genus and species barriers.
Abbreviations
- BT:
-
Bluetongue
- BTV:
-
Bluetongue virus
- CDC:
-
Center for Disease Control
- PR:
-
Prevalence ratio
- SBV:
-
Schmallenberg virus
- wsp :
-
Wolbachia surface protein gene
References
Nolan DV, Dallas JF, Piertney SB, Mordue Luntz AJ. Incursion and range expansion in the bluetongue vector Culicoides imicola in the Mediterranean basin: a phylogeographic analysis. Med Vet Entomol. 2008;22:340–51.
Tago D, Hammitt JK, Thomas A, Raboisson D. Cost assessment of the movement restriction policy in France during the 2006 bluetongue virus episode (BTV-8). Prev Vet Med. 2014;117:577–89.
Velthuis AG, Saatkamp HW, Mourits MC, de Koeijer AA, Elbers AR. Financial consequences of the Dutch bluetongue serotype 8 epidemics of 2006 and 2007. Prev Vet Med. 2010;93:294–304.
Mellor PS, Boorman J, Baylis M. Culicoides biting midges: their role as arbovirus vectors. Annu Rev Entomol. 2000;45:307–40.
Balenghien T, Pages N, Goffredo M, Carpenter S, Augot D, Jacquier E, et al. The emergence of Schmallenberg virus across Culicoides communities and ecosystems in Europe. Prev Vet Med. 2014;116:360–9.
Pagès N, Talavera S, Verdún M, Pujol N, Valle M, Bensaid A, Pujols J. Schmallenberg virus detection in Culicoides biting midges in Spain: First laboratory evidence for highly efficient infection of Culicoides of the Obsoletus complex and Culicoides imicola. Transbound Emerg Dis 2017;00:1–6.
Carpenter S, Mellor PS, Torr SJ. Control techniques for Culicoides biting midges and their application in the U.K. and northwestern Palaearctic. Med Vet Entomol. 2008;22:175–87.
Maclachlan NJ, Mayo CE. Potential strategies for control of bluetongue, a globally emerging, Culicoides-transmitted viral disease of ruminant livestock and wildlife. Antivir Res. 2013;99:79–90.
Feldhaar H, Gross R. Insects as hosts for mutualistic bacteria. Int J Med Microbiol. 2009;299:1–8.
Floate KD, Kyei-Poku GK, Goghlin PC. Overview and relevance of Wolbachia bacteriain biocontrol research. Biocontrol Sci Tech. 2006;16:767–88.
Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–20.
Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–51.
Stouthamer R, Breeuwer JA, Hurst GD. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol. 1999;53:71–102.
Zchori-Fein E, Perlman SJ. Distribution of the bacterial symbiont Cardinium in arthropods. Mol Ecol. 2004;13:2009–16.
Caragata EP, Dutra HL, Moreira LA. Exploiting intimate relationships: controlling mosquito-transmitted disease with Wolbachia. Trends Parasitol. 2016;32:207–18.
Medlock J, Atkins KE, Thomas DN, Aksoy S, Galvani AP. Evaluating paratransgenesis as a potential control strategy for African trypanosomiasis. PLoS Negl Trop Dis. 2013;7:e2374.
Nakamura Y, Kawai S, Yukuhiro F, Ito S, Gotoh T, Kisimoto R, et al. Prevalence of Cardinium bacteria in planthoppers and spider mites and taxonomic revision of “Candidatus Cardinium hertigii” based on detection of a new Cardinium group from biting midges. Appl Environ Microbiol. 2009;75:6757–63.
Morag N, Klement E, Saroya Y, Lensky I, Gottlieb Y. Prevalence of the symbiont Cardinium in Culicoides (Diptera: Ceratopogonidae) vector species is associated with land surface temperature. FASEB J. 2012;26:4025–34.
Lewis SE, Rice A, Hurst GD, Baylis M. First detection of endosymbiotic bacteria in biting midges Culicoides pulicaris and Culicoides punctatus, important Palaearctic vectors of bluetongue virus. Med Vet Entomol. 2014;28:453–6.
Mee PT, Weeks AR, Walker PJ, Hoffmann AA, Duchemin JB. Detection of low-level Cardinium and Wolbachia infections in Culicoides. Appl Environ Microbiol. 2015;81:6177–88.
Muñoz-Muñoz F, Ramoneda J, Pagès N, Pujol N, Talavera S. Is the morphology of Culicoides intersexes parasitized by mermithid nematodes a parasite adaptation? A morphometric approach to Culicoides circumscriptus (Diptera: Ceratopogonidae). J Invertebr Pathol. 2016;135:1–9.
McMeniman CJ, Lane RV, Cass BN, Fong AW, Sidhu M, Wang YF, et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323:141–4.
Bian G, Joshi D, Dong Y, Lu P, Zhou G, Pan X, et al. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340:748–51.
Sinkins SP, Godfray HC. Use of Wolbachia to drive nuclear transgenes through insect populations. Proc Biol Sci. 2004;271:1421–6.
Ruang-Areerate T, Kittayapong P. Wolbachia transinfection in Aedes aegypti: a potential gene driver of dengue vectors. Proc Natl Acad Sci USA.2006;103:12534–9.
Delécolle JC. Nouvelle contribution à l'étude systématique et iconographique des espèces du genre Culicoides (Diptera: Ceratopogonidae) du Nord-Est de la France. Strasbourg: PhD Thesis, Université Louis Pasteur du Strasbourg; 1985.
QGIS Development Team, 2016. QGIS geographic information system. Open Source Geospatial Foundation Project. http://www.qgis.org/
Duron O, Bouchon D, Boutin S, Bellamy L, Zhou L, Engelstadter J, et al. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 2008;6:27.
Pagès N, Sarto IMV. Differentiation of Culicoides obsoletus and Culicoides scoticus (Diptera: Ceratopogonidae) based on mitochondrial cytochrome oxidase subunit I. J Med Entomol. 2005;42:1026–34.
Nolan DV, Carpenter S, Barber J, Mellor PS, Dallas JF, Mordue Luntz AJ, et al. Rapid diagnostic PCR assays for members of the Culicoides obsoletus and Culicoides pulicaris species complexes, implicated vectors of bluetongue virus in Europe. Vet Microbiol. 2007;124:82–94.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–8.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Morag N, Mullens BA, Gottlieb Y. Assessment of survival and body size variation of Culicoides imicola (Diptera: Ceratopogonidae) as functions of “Candidatus Cardinium” (Bacteroidetes) infection status. Appl Environ Microbiol. 2013;79:6260–3.
Ritchie SA, Townsend M, Paton CJ, Callahan AG, Hoffmann AA. Application of wMelPop Wolbachia strain to crash local populations of Aedes aegypti. PLoS Negl Trop Dis. 2015;9:e0003930.
Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–3.
Aliota MT, Walker EC, Uribe Yepes A, Dario Velez I, Christensen BM, Osorio JE. The wMel strain of Wolbachia reduces transmission of chikungunya virus in Aedes aegypti. PLoS Negl Trop Dis. 2016;10:e0004677.
Raychoudhury R, Baldo L, Oliveira DCSG, Werren JH. Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution. 2009;63:165–83.
Bourtzis K. Wolbachia-based technologies for insect pest population control. Adv Exp Med Biol. 2008;627:104–13.
Acknowledgements
The authors would like to thank Departament d’Agricultura, Ramaderia, Pesca i Alimentació of Generalitat de Catalunya for logistic support. We would like to thank the owners of farms for permission to place the traps and people involved in sample collection.
Funding
National grant FAU2008–0019 partially funded this study from Instituto Nacional de Investigaciones Agrarias (INIA).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files. The sequences are submitted in the GenBank database under the accession numbers MF179641–MF179663 (Wolbachia) and MF188893–MF188900 (Cardinium).
Author information
Authors and Affiliations
Contributions
NPa conceived and designed the study. NPa, FM and ST contributed to Culicoides field sampling. FM, MV and ST performed morphological identification of Culicoides. NPa, NPu performed genetic analyses. NPa drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: Table S1.
NCBI published sequences used to better reconstruct the evolutionary relationship of Wolbachia and Cardinium isolates. (DOCX 48 kb)
Additional file 2: Table S2.
Diagnostic tests performed in Culicoides to detect Wolbachia and Cardinium endosymbionts. Infection frequencies are shown in parentheses for populations where endosymbionts were detected in Culicoides. (DOCX 47 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Pagès, N., Muñoz-Muñoz, F., Verdún, M. et al. First detection of Wolbachia-infected Culicoides (Diptera: Ceratopogonidae) in Europe: Wolbachia and Cardinium infection across Culicoides communities revealed in Spain. Parasites Vectors 10, 582 (2017). https://doi.org/10.1186/s13071-017-2486-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-017-2486-9