Abstract
Background
Toxoplasma gondii is a widespread protozoan parasite that infects humans and other animals. Previous studies indicate some genotypes of T. gondii are more frequently isolated in wildlife than agricultural animals, suggesting a wild/feral animal diversity model. To determine seroprevalence and genetic diversity of T. gondii in southeastern US wildlife, we collected sera from 471 wild animals, including 453 mammals and 18 birds, between 2011 and 2014. These serum samples were assayed for T. gondii infection using the modified agglutination test (MAT). Heart or tongue tissues from 66 seropositive animals were bioassayed in mice and 19 isolates were obtained. The isolated parasites were genotyped by the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method employing 10 genetic markers.
Results
One hundred and ninety-six of 471 samples (41.6%) had a titer ≥1:32 and were considered positive for T. gondii infection. Of 453 mammals, 195 (43%) were seropositive, whereas only one (5.6%) of 18 birds was seropositive. The seroprevalence in mammals was significantly higher than in the birds. Mammalian hosts with adequate samples size (≥ 20) comprised white-tailed deer (n = 241), feral hogs (n = 100), raccoons (n = 34) and coyotes (n = 22), with seroprevalences of 41.0%, 51.0%, 50.0% and 72.7%, respectively. Coyotes had significantly higher seroprevalence than the white-tailed deer. Genotyping revealed five distinct genotypes, including the ToxoDB PCR-RFLP genotype #5 (a.k.a type 12) for 15 isolates, genotype #3 (a.k.a. type II) for 1 isolate, and genotypes #154, #167 and #216, each for 1 isolate. The results showed moderate to high infection rates of T. gondii in white-tailed deer, feral hogs, raccoons and coyotes. Genotyping results indicated limited genetic diversity and a dominance of genotype #5, which has been reported as a major type in wildlife in North America.
Conclusions
We conclude that T. gondii infection is common in game animals (white-tailed deer and feral hogs) in the southeastern US, which may pose a food safety risk to humans. Further research is necessary to understand T. gondii transmission from wildlife to farm animals and humans.
Similar content being viewed by others
Background
Toxoplasmosis, caused by Toxoplasma gondii, is zoonotic and considered a leading cause of human morbidity attributed to food borne illness in the United States [1], and it is estimated that one-third of the world’s population is infected by this pathogen [2]. Women infected with T. gondii during pregnancy can have variable consequences including pregnancy complications, stillbirths and abortions. In immunocompromised patients, such as those with AIDS, encephalitis may occur, which is often fatal [2]. Toxoplasmosis is one of five neglected parasitic infections that have been targeted by the Centers for Disease Control and Prevention (CDC) for public health action. Infection with T. gondii can occur by ingestion of microscopic oocysts in contaminated food or water, or by ingestion of tissue cysts in undercooked or raw meat [2, 3], making it an important foodborne zoonotic pathogens.
Toxoplasma gondii infection occurs in many species of wild mammals and birds, particularly those that are carnivorous or ground dwelling. Clinical toxoplasmosis occurs in a wide variety of US wildlife, including threatened and endangered terrestrial and marine mammals and birds [4, 5]. Epidemiology studies of white-tailed deer populations have reported seroprevalence from 30% to 76% in areas including Pennsylvania, Minnesota, Mississippi, New Jersey, Iowa and Ohio [6,7,8,9,10]. A range of seroprevalence (15–84%) was observed in raccoons from Iowa, New Jersey, Ohio, Kansas, Illinois, Florida, Pennsylvania, Virginia and Wisconsin [11,12,13,14]. A high seroprevalence in red and gray foxes (85.9%) was reported in Kentucky, Indiana, Michigan and Ohio [15, 16] and wild hogs from California and black bears from Pennsylvania also show seroprevalence of 17% and 75–80%, respectively [8, 17]. Antibodies against T. gondii (7–17%) were detected in wolves from remote areas in Alaska [18, 19]. Genotyping of wildlife isolates suggests that wild animals maintain a much greater diversity of T. gondii genotypes than agricultural animals [20,21,22]. There is no reported association between T. gondii genotypes and disease manifestation, but some evidence suggests a relationship. For example, in South America, where wild animal populations are more dominant, severe cases of human toxoplasmosis were reported even in immunocompetent adults [23,24,25,26], and the majority of these infections were attributed to unique genotypes. Recent studies have reported the presence of numerous genotypes in wildlife populations in North America. Currently, ToxoDB PCR-RFLP genotypes #4 and #5, also known as type 12, are recognized as the dominant type in North America wildlife [20, 21]. It is likely that some of these T. gondii strains from wildlife are highly virulent, posing a potential wildlife health risk and a higher risk for severe toxoplasmosis if transmitted in human populations.
The role of wildlife in the transmission of T. gondii demands increased efforts to catalog the major sources of human T. gondii infection. Continued characterization is critical to understanding the potential risks of T. gondii to wildlife populations and its zoonotic implications. Seroprevalence and genotyping data from the southeast region of the United States have been insufficient to determine the pattern of T. gondii transmission in the area. Hence, in this study, we focused on determining seroprevalence and characterizing strains isolated from wildlife in this region.
Methods
Serum with or without corresponding fresh heart or tongue tissue samples was collected from hunter-killed, road killed, nuisance killed (i.e. feral hogs), or research collected animals from multiple southeastern states (Table 1). Tissue samples were refrigerated until serological screening was completed.
Screening for T. gondii was performed at the clinical parasitology laboratory at the University of Tennessee, College of Veterinary Medicine using the MAT test as previously described [27, 28]. This assay is used to detect anti-T. gondii antibodies in blood, serum and other bodily fluids from a wide variety of wildlife and domestic species. Animals were considered Toxoplasma positive if IgG antibodies were detected at ≥ 1:32 dilution on MAT. Three to 5 g of heart or tongue tissue from some seropositive hosts were processed and used in bioassays of mice to propagate T. gondii [29]. To facilitate isolation of T. gondii, mice were treated with 15 μl/ml dexamethasone in drinking water at the time of inoculation of processed animal tissues. Mice showing clinical signs of infection (roughed fur and lethargic) were terminated, peritoneal lavage are collected and inoculated to cell culture to expand the parasites. All nonclinical mice were terminated on day 14 post-inoculation, peritoneal lavage was collected and inoculated to cell culture. Isolated T. gondii strains were genotyped by multiplex multilocus nested PCR-RFLP (Mn-PCR-RFLP) employing 10 genetic markers [30].
To compare seroprevalence of different populations, data analysis was performed using statistical software SAS version 9.4. Chi-square tests were conducted to determine if there was statistically significant difference among different sampling groups. Logistic regression was used to compute the odds ratios of infection among different groups. Association of serum MAT titer with success of isolating T. gondii in bioassay was assessed by linear regression analysis using SAS GLM procedure (SAS 9.4).
Results
Seroprevalence of T. gondii
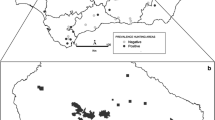
A total of 471 serum/plasma samples were collected from 31 wildlife species (16 mammal and 15 bird species) between 2011 and 2014 (Table 2). Samples originated in six southeast states, comprising Alabama, Georgia, Kentucky, North Carolina, South Carolina and Tennessee (Table 1). From 471 samples, 41.6% (196/471) had MAT titers ≥ 1:32 and were considered positive for T. gondii infection (Tables 1, 2). Nine mammalian (white-tailed deer, opossum, raccoon, coyote, feral hog, woodchuck, elk, gray fox and mink) and 1 bird species (rock pigeon) collected from five southeastern states had seropositive individuals (Tables 1, 2). Mammal hosts with samples size ≥ 10 individuals comprised white-tailed deer (n = 241), feral hogs (n = 100), raccoons (n = 34), coyotes (n = 22), opossum (n = 12) and gray squirrels (n = 14) and had seroprevalences of 41%, 51%, 50%, 72.7%, 50% and 0%, respectively.
In Tennessee, 309 serum samples from 29 animal species were collected and tested from 10 counties/sites (Table 1). Overall, 37.5% (116/309) were positive for T. gondii infection. In South Carolina, 74 serum samples from white-tailed deer in Laurens County were tested, with 44.5% (33/74) seropositive. In North Carolina, 74 serum samples (73 from feral hogs, 1 from a black bear) were collected from the GSMNP (Table 1), 54.1% (40/74) positive. For feral hogs, 54.8% (40/73) were positive to T. gondii infection. In Georgia, 6 serum samples were collected from 5 coyotes and 1 gray fox in Jefferson and Putnam counties (Table 1). The 5 samples from coyotes in Putnam County were all seropositive. Four serum samples from 4 animal species in Alabama were all negative (Table 1). Two of 4 samples from elk in Kentucky were positive (50%).
Comparison of seroprevalence in different wildlife hosts and geographical locations
Seroprevalence in mammals was 39.2% (195/453), which was significantly higher than in birds (5.6%, 1/18) (Chi-square test: χ 2 = 6.10, df = 1, P = 0.014; Odds ratio: 12.84; 95% CI: 1.695–97.26). Among the mammal populations with sample size ≥ 20, including Tennessee (115/293), South Carolina (33/74) and North Carolina (40/74), there was no statistically significant difference in seroprevalence (Chi-square test: χ 2 = 5.36, df = 2, P = 0.068). Comparison of seroprevalence for white-tailed deer and feral hogs that had sample size ≥ 20 in different geographical locations was performed. Seroprevalence rates in white-tailed deer from Ames Plantation (Tennessee), Oak Ridge (Tennessee) and Laurens (South Carolina) were 51.9, 28.1 and 44.6%, respectively (Table 1). White-tailed deer from Ames Plantation and Laurens had significantly higher odds of being positive than those in Oak Ridge (Chi-square test: χ 2 = 8.14, df = 2, P = 0.017), with Laurens vs Oak Ridge, odds ratio 2.057 (95% CI: 1.009–4.192); Ames Plantation vs Oak Ridge, odds ratio 2.763 (95% CI: 1.365–5.590). Seroprevalence rates in white-tailed deer from Ames Plantation and Laurens were not significantly different (odds ratio 1.343, 95% CI: 0.708–2.548). Seroprevalence rates in feral hogs from GSMNP Tennessee and GSMNP North Carolina were 40.7 and 54.8%, respectively (Table 1). There was no statistically significant difference between the two groups (Chi-square test: χ 2 = 1.54, df = 1, P = 0.215; Odds ratio: 1.763; 95% CI: 0.720–4.317).
Comparison of seroprevalence was also conducted for wildlife species that had sample size ≥ 20 regardless of geographical locations. These species included white-tailed deer (n = 241), feral hogs (n = 100), raccoons (n = 34) and coyotes (n = 22), which had seroprevalence rates of 41.0, 51.0, 50.0, and 72.7%, respectively (Table 2). Significant difference was detected (Chi-square test: χ 2 = 9.24, df = 3, P = 0.026), with coyotes having a significantly higher infection rate than white-tailed deer (odds ratio 3.825, 95% CI 1.446–10.117). No differences were detected among other species.
Isolation and genotyping of T. gondii strains
Tissue (hearts and tongues) from 66 seropositive wildlife samples were bioassayed in mice. These samples comprised: 33 from white-tailed deer, 11 from feral hogs, 8 from raccoons, 8 from coyotes, 2 from elks, 2 from opossums, 1 from mink and 1 from gray fox. Nineteen T. gondii isolates were obtained by bioassay (13 from white-tailed deer, 3 from feral hogs, 2 from coyotes and 1 from a mink) (Table 3). For tissue samples with MAT titers of 32, 128, 512, 2048, 4096 and ≥ 8192, the rates of obtaining T. gondii isolates in bioassay were 0, 15, 12.5, 20, 66.7 and 62.5%, respectively. There was a significant correlation between MAT titers and the success rates of bioassay (GLM linear regression coefficient r = 0.88, P = 0.021).
The 19 T. gondii isolates were genotyped by the 10 PCR-RFLP markers (Table 4). Five distinct genotypes were identified: ToxoDB PCR-RFLP genotype #5 (15 isolates), #3 (1 isolate), #154 (1 isolate), #167 (1 isolate) and #216 (1 isolate). Of the 13 isolates obtained from white-tailed deer, 9 were from South Carolina and 4 from Tennessee.
Discussion
The present study demonstrates that T. gondii infection is widespread in wild mammals from the southeastern United States. We collected sera from 471 wild animals (453 mammals and 18 birds) between 2011 and 2014. Overall, 41.6% were positive for T. gondii infection, however, only one of the 18 birds was seropositive (Table 2). The seroprevalence in mammals was significantly higher than in the birds. Among the most frequently sampled mammal species (white-tailed deer, feral hog, raccoon and coyote, n ≥ 20 each), seroprevalence varies from 41% to 72.7%, with that for coyote significantly higher than for white-tailed deer (Table 2), which supports the general idea that carnivores have higher infection rates than herbivores.
Among the three geographical locations with mammal samples size ≥ 20, Tennessee (n = 293), South Carolina (n = 74) and North Carolina (n = 74), seroprevalence rates varied from 39.2% to 54.1%, however, there was no statistically significant difference. Among white-tailed deer populations from three different locations, Ames Plantation (Tennessee), Oak Ridge (Tennessee), and Laurens (South Carolina), seroprevalence in Oak Ridge was significantly lower than the other two populations, which warrants future studies to understand what factors contribute to such a difference. Seroprevalence rates in feral hogs from North Carolina and Tennessee sides of the Great Smoky Mountains National Park were, in general, not significantly different, which is expected given the similar environment.
In this study, the success rate of bioassay was 28.8% (19/66). Efficiency of bioassay can be affected by many factors, such as how long the tissue samples were stored before inoculated to mice, the amount of tissues used, and the type of tissues used. In addition, tissue cysts may not evenly distribute in the muscle or brain tissues of infected animals, and successfully obtaining cysts variable between samples. We did an analysis of MAT titers vs success rates in bioassay; it showed a positive correlation, suggesting higher titers may have higher parasite load in the tissues.
Genotype #5 (a.k.a. type 12) is the most common circulating genotype in wildlife in this region of the US, which is in agreement with previous studies reporting the prevalence of genotype #5 in white-tailed deer populations [20, 31]. Genotypes #156 and #167 have been previously reported from goats in the USA [32]. Two isolates from coyote (TgWtdTn122 and TgWtdTn142) and 1 mink isolate (TgMnkTn17) obtained from Tennessee, belong to genotype #5 and #3, respectively. Genotype #3 (type II) of T. gondii is the most dominant lineage distributed globally. Furthermore, the 2 feral hog isolates (TgHogNc194 and TgHogNc227) from North Carolina also belonged to genotype #5, which is commonly distributed in North America [31]. We were unable to assess the virulence of T. gondii strains in mice during the bioassay, as mice were treated with dexamethasone to suppress their immune responses and the experiments were terminated on day 14 post-infection.
Conclusions
In addition to the commonly observed genotypes, we also isolated several non-clonal types circulating in sampled populations. This is of interest, as previous epidemiological studies have reported a link between the prevalence of non-clonal genotypes and cases of congenital ocular and severe disseminated toxoplasmosis in areas such as Brazil [33]. White-tailed deer is one of the dominant wildlife species found in North America and venison a common game meat. Thus, the high seroprevalence in this species indicates that deer could serve as a potential source of human infection. Hence, people consuming wild venison should be advised to cook the meat properly and use caution while handling the raw meat. Future genotyping and seroprevalence studies in wildlife hosts, and analysis of their role in the transmission cycle, will increase the understanding of risks associated with T. gondii in human populations.
Abbreviations
- MAT:
-
Modified agglutination test
- PCR-RFLP:
-
Polymerase chain reaction-restriction fragment length polymorphism
References
Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, et al. Foodborne illness acquired in the United States - major pathogens. Emerg Infect Dis. 2011;17:7–15.
Dubey JP. Toxoplasmosis of animals and humans. 2nd ed. Boca Raton: Taylor & Francis; 2010.
Tenter AM, Heckeroth AR, Weiss LM. Toxoplasma gondii: from animals to humans. Int J Parasitol. 2000;30:1217–58.
Work TM, Massey JG, Rideout BA, Gardiner CH, Ledig DB, Kwok OCH, et al. Fatal toxoplasmosis in free-ranging endangered ‘Alala from Hawaii. J Wildl Dis. 2000;36:205–12.
Smith LN, Waltzek TB, Rotstein DS, Francis-Floyd R, Walsh MT, Wellehan JF, et al. Disseminated toxoplasmosis Toxoplasma gondii in a wild Florida manatee Trichechus manatus latirostris and seroprevalence in two wild populations. Dis Aquat Org. 2016;122:77–83.
Ballash GA, Dubey JP, Kwok OC, Shoben AB, Robison TL, Kraft TJ, Dennis PM. Seroprevalence of Toxoplasma gondii in white-tailed deer (Odocoileus virginianus) and free-roaming cats (Felis catus) across a suburban to urban gradient in northeastern Ohio. EcoHealth. 2015;12:359–67.
Dubey JP, Dennis PM, Verma SK, Choudhary S, Ferreira LR, Oliveira S, et al. Epidemiology of toxoplasmosis in white tailed deer (Odocoileus virginianus): occurrence, congenital transmission, correlates of infection, isolation, and genetic characterization of Toxoplasma gondii. Vet Parasitol. 2014;202:270–5.
Dubey JP, Jones JL. Toxoplasma gondii infection in humans and animals in the United States. Int J Parasitol. 2008;38:1257–78.
Humphreys JG, Stewart RL, Dubey JP. Prevalence of Toxoplasma gondii antibodies in sera of hunter-killed white-tailed deer in Pennsylvania. Am J Vet Res. 1995;56:172–3.
Vanek JA, Dubey JP, Thulliez P, Riggs MR, Stromberg BE. Prevalence of Toxoplasma gondii antibodies in hunter-killed white-tailed deer (Odocoileus virginianus) in four regions of Minnesota. J Parasitol. 1996;82:41–4.
Brillhart DB, Fox LB, Dubey JP, Upton SJ. Seroprevalence of Toxoplamsa gondii in wild animals in Kansas. J Helminthol Soc Wash. 1994;61:117–21.
Dubey JP, Hamir AN, Hanlon CA, Rupprecht CE. Prevalence of Toxoplasma gondii infection in raccoons. J Am Vet Med Assoc. 1992;200:534–6.
Hill RE Jr, Zimmerman JJ, Wills RW, Patton S, Clark WR. Seroprevalence of antibodies against Toxoplasma gondii in free-ranging mammals in Iowa. J Wildl Dis. 1998;34:811–5.
Mitchell MA, Hungeford LL, Nixon C, Esker T, Sullivan J, Koerkenmeier R, Dubey JP. Serologic survey for selected infectious disease agents in raccoons from Illinois. J Wildl Dis. 1999;35:347–55.
Dubey JP, Graham DH, De Young RW, Dahl E, Eberhard ML, Nace EK, et al. Molecular and biologic characteristics of Toxoplasma gondii isolates from wildlife in the United States. J Parasitol. 2004;90:67–71.
Walton BC, Walls KW. Prevalence of toxoplasmosis in wild animals from Fort Stewart, Georgia, as indicated by serological tests and mouse inoculation. Am J Trop Med Hyg. 1964;13:530–3.
Clark RK, Jessup DA, Hird DW, Ruppanner R, Meyer ME. Serologic survey of California wild hogs for antibodies against selected zoonotic disease agents. J Am Vet Med Assoc. 1983;183:1248–51.
Stieve E, Beckmen K, Kania SA, Widner A, Patton S. Neospora caninum and Toxoplasma gondii antibody prevalence in Alaska wildlife. J Wildl Dis. 2010;46:348–55.
Zarnke RL, Dubey JP, Kwok OC, Ver Hoef JM. Serologic survey for Toxoplasma gondii in selected wildlife species from Alaska. J Wildl Dis. 2000;36:219–24.
Dubey JP, Velmurugan GV, Rajendran C, Yabsley MJ, Thomas NJ, Beckmen KB, et al. Genetic characterisation of Toxoplasma gondii in wildlife from North America revealed widespread and high prevalence of the fourth clonal type. Int J Parasitol. 2011;41:1139–47.
Khan A, Dubey JP, Su C, Ajioka JW, Rosenthal BM, Sibley LD. Genetic analyses of atypical Toxoplasma gondii strains reveal a fourth clonal lineage in North America. Int J Parasitol. 2011;41:645–55.
Velmurugan GV, Su C, Dubey JP. Isolate designation and characterization of Toxoplasma gondii isolates from pigs in the United States. J Parasitol. 2009;95:95–9.
Darde ML, Villena I, Pinon JM, Beguinot I. Severe toxoplasmosis caused by a Toxoplasma gondii strain with a new isoenzyme type acquired in French Guyana. J Clin Microbiol. 1998;36:324.
Demar M, Ajzenberg D, Maubon D, Djossou F, Panchoe D, Punwasi W, et al. Fatal outbreak of human toxoplasmosis along the Maroni River: epidemiological, clinical, and parasitological aspects. Clin Infect Dis. 2007;45:88–95.
Demar M, Hommel D, Djossou F, Peneau C, Boukhari R, Louvel D, et al. Acute toxoplasmosis in immunocompetent patients hospitalized in an intensive care unit in French Guiana. Clin Microbiol Infect. 2012;18:221–31.
Gilbert RE, Freeman K, Lago EG, Bahia-Oliveira LM, Tan HK, Wallon M, et al. Ocular sequelae of congenital toxoplasmosis in Brazil compared with Europe. PLoS Negl Trop Dis. 2008;2:277.
Dubey JP, Desmonts G. Serological responses of equids fed Toxoplasma gondii oocysts. Equine Vet J. 1987;19:337–9.
Desmonts G, Remington JS. Direct agglutination test for diagnosis of Toxoplasma infection: method for increasing sensitivity and specificity. J Clin Microbiol. 1980;11:562–8.
Dubey JP. Refinement of pepsin digestion method for isolation of Toxoplasma gondii from infected tissues. Vet Parasitol. 1998;74:75–7.
Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology. 2010;137:1–11.
Shwab EK, Zhu XQ, Majumdar D, Pena HF, Gennari SM, Dubey JP, et al. Geographical patterns of Toxoplasma gondii genetic diversity revealed by multilocus PCR-RFLP genotyping. Parasitology. 2014;141:453–61.
Dubey JP, Rajendran C, Ferreira LR, Martins J, Kwok OC, Hill DE, et al. High prevalence and genotypes of Toxoplasma gondii isolated from goats, from a retail meat store, destined for human consumption in the USA. Int J Parasitol. 2011;41:827–33.
Holland GN. Ocular toxoplasmosis: a global reassessment. Part I: epidemiology and course of disease. Am J Ophthalmol. 2003;136:973–88.
Acknowledgements
We acknowledge the following that assisted on the project: Sharon Patton, Heidi Wyrosdick, Caroline Grunewald, Lauren Maestas, Teresa Moody, Al Claiborne, Lauren Henderson, Caroline Brown, and Jill Wilson. Julia Zhu assisted with statistical analysis of the data.
Funding
Funding for this project was provided by the University of Tennessee’s MCERV Program 2011 to RWG and CS.
Availability of data and materials
All data generated and analyzed in this study are included in this paper.
Author information
Authors and Affiliations
Contributions
RWG and CS: participated in experimental design and coordination of the study. RWG and PS: participated in drafting and revision of the manuscript. GH, WHS, AH and MS: participated in sample collection and revision of the manuscript. AC: contributed to serological analysis of samples and revision of the manuscript. XZ, PS and CS: contributed to molecular genetic analysis of samples. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The use and care of laboratory mice for this study were approved by the Institutional Animal Care and Use Committee of the University of Tennessee (Permit ID Number: 1419-0111).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Gerhold, R.W., Saraf, P., Chapman, A. et al. Toxoplasma gondii seroprevalence and genotype diversity in select wildlife species from the southeastern United States. Parasites Vectors 10, 508 (2017). https://doi.org/10.1186/s13071-017-2456-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-017-2456-2