Abstract
Trypanosomatids are protozoan parasites of the class Kinetoplastida predominately restricted to invertebrate hosts (i.e. possess a monoxenous life-cycle). However, several genera are pathogenic to humans, animals and plants, and have an invertebrate vector that facilitates their transmission (i.e. possess a dixenous life-cycle). Phytomonas is one dixenous genus that includes several plant pathogens transmitted by phytophagous insects. Trypanosoma and Leishmania are dixenous genera that infect vertebrates, including humans, and are transmitted by hematophagous invertebrates. Traditionally, monoxenous trypanosomatids such as Leptomonas were distinguished from morphologically similar dixenous species based on their restriction to an invertebrate host. Nonetheless, this criterion is somewhat flawed as exemplified by Leptomonas seymouri which reportedly infects vertebrates opportunistically. Similarly, Novymonas and Zelonia are presumably monoxenous genera yet sit comfortably in the dixenous clade occupied by Leishmania. The isolation of Leishmania macropodum from a biting midge (Forcipomyia spp.) rather than a phlebotomine sand fly calls into question the exclusivity of the Leishmania-sand fly relationship, and its suitability for defining the Leishmania genus. It is now accepted that classic genus-defining characteristics based on parasite morphology and host range are insufficient to form the sole basis of trypanosomatid taxonomy as this has led to several instances of paraphyly. While improvements have been made, resolution of evolutionary relationships within the Trypanosomatidae is confounded by our incomplete knowledge of its true diversity. The known trypanosomatids probably represent a fraction of those that exist and isolation of new species will help resolve relationships in this group with greater accuracy. This review incites a dialogue on how our understanding of the relationships between certain trypanosomatids has shifted, and discusses new knowledge that informs the present taxonomy of these important parasites.
Similar content being viewed by others
Background
The Trypanosomatidae are a diverse family of protozoan parasites that are predominately monoxenous. Nonetheless, some trypanosomatids occupy a dixenous (syn. digenetic) niche [1]. Phytomonas is one example of a dixenous trypanosomatid genus, transmitted by phytophagous insects and parasitising a variety of plants. However, the trypanosomatids were brought to prominence by two genera, Trypanosoma and Leishmania, attributable to their role as human pathogens. Trypanosoma and Leishmania are obligatorily dixenous, possess zoonotic or anthroponotic life-cycles, and are transmitted by hematophagous insects. These parasites cause devastating human diseases including Human African Trypanosomiasis (aetiology: Trypanosoma brucei), Chagas disease (aetiology: Trypanosoma cruzi), and the leishmaniases, which are attributable to roughly 20 species of Leishmania [2,3,4,5,6,7,8] (Table 1). These diseases are associated with a spectrum of symptoms differing in severity and mechanisms of pathogenesis [8]. These dixenous species also differ in their preferred site of development within their vertebrate hosts which influences disease phenotype and severity [9,10,11]. With the exception of the gambiense form of Human African Trypanosomiasis (HAT) and infections caused by Leishmania tropica and Leishmania donovani, for which humans are considered the main reservoir, trypanosomatid-associated diseases are predominately zoonotic, with animal reservoirs playing a key role in maintaining endemicity [8, 12].
Trypanosoma spp. are ubiquitous, infecting almost all vertebrate classes [13], with vectors ranging from leeches, to biting flies and bugs [8, 14]. The tsetse fly (genus: Glossina) and bugs of the Triatominae subfamily (i.e. “kissing” bugs) are the natural vectors of Trypanosoma brucei and Trypanosoma cruzi, respectively [8] (Fig. 1). Human African Trypanosomiasis is endemic in 36 sub-Saharan countries with studies estimating that 61 million people are at risk of contracting the disease through the bite of an infected tsetse fly [15]. Domestic and wild animals serve as reservoirs for T. brucei infection, with the reported human prevalence varying between communities [2]. Between 2004 and 2014, the number of new cases reported has significantly dropped from 17,616 to only 3,796 with the total number of estimated annual cases plummeting from 50,000–70,000 to less than 15,000 [15]. Chagas disease is endemic in Central and South America, with an estimated 8 million individuals infected [16]. Humans are an incidental host of Trypanosoma cruzi, which is a zoonotic parasite predominantly infecting native wildlife, domestic dogs and cats, which are reservoirs of Chagas disease [17].
Vectors and invertebrate hosts of some trypanosomatids. a A female Phlebotomus sp. sand fly which is a vector of Leishmania spp. Citation: Hailu et al. Visceral leishmaniasis: New health tools are needed. PLoS Med. 2005;2(7):590–594 [151]. b A female Simulium (Morops) dycei, which is the host of Zelonia australiensis. Citation: Barratt et al. Isolation of novel trypanosomatid, Zelonia australiensis sp. nov. (Kinetoplastida: Trypanosomatidae) provides support for a Gondwanan origin of dixenous parasitism in the Leishmaniinae. PLOS Negl Trop Dis. 2017;11(1):e0005215 [68]. c The tsetse fly is the vector of Trypanosoma brucei; the aetiological agent of Human African Trypanosomiasis (http://researchnews.plos.org/2016/08/08/under-my-skin/) [152]. d A Triatomine “kissing” bug, which is the natural vector of Trypanosoma cruzi; the aetiological agent of Chagas disease. Citation: Curtis-Robles et al. Combining public health education and disease ecology research: using citizen science to assess Chagas disease entomological risk in Texas. PLoS Neglect Trop Dis. 2015;9(12):12 [153]. a, b, d Copyright: Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/)
Female phlebotomine sand flies (Diptera: Psychodidae: Phlebotominae) are natural vectors for Leishmania transmission [18, 19], and roughly 70 animal species serve as reservoirs for human pathogenic Leishmania species, including rodents [20], dogs [21] and other mammals [22]. Australia and Antarctica had long been considered the only continents free of endemic Leishmania though the discovery of an Australian macropod-infecting species and its midge vector, Forcipomyia (Lasiohelea) spp. [20, 22, 23], overturned this and called into question the exclusivity of the sand fly-Leishmania interaction.
Compared to their dixenous cousins, the life-cycle and habits of monoxenous trypanosomatids are obscure. By 2001, monoxenous trypanosomatids had been identified from roughly 350 insect species only, while more than 900 vertebrate hosts had been identified for the dixenous genera which are far fewer in number [24]. Due to their limited impact on human and animal health, the monoxenous trypanosomatids have received little attention from parasitologists historically. Despite this, interest in the monoxenous species has revived in recent years [25,26,27,28]. From a taxonomic perspective, trypanosomatids are now amongst the most extensively studied protozoans, as reflected by the recent surge in publications on this topic that form the basis of the trypanosomatid taxonomic system [26, 27, 29].
Trypanosomatid systematics was traditionally based on host preferences and specialised life-cycle stages, characterised by the presence or absence of several defined morphotypes (Fig. 2) [30,31,32]. Under the classical system of trypanosomatid taxonomy, the various Leishmania species were differentiated by their respective specialised proliferative stages [18]. Advances in molecular biology revolutionised the field by providing genetic evidence for true ancestral relationships [27, 33]. As a consequence, major flaws were identified in the classical system which underwent a series of major revisions [26,27,28,29]. However, the field would continue to benefit from studies aiming to isolate new species. Of the estimated one million known insect species described, only 2,500 of these have been studied closely for the presence of trypanosomatids [24, 26, 29, 34]. The isolation and molecular characterisation of new species will help to decipher relationships between trypanosomatids with greater resolution [24].
The six major morphotype classes of trypanosomatids. a Trypomastigote. b Epimastigote. c Amastigote. d Opisthomastigote. e Choanomastigote. f Promastigote. Forms a and b represent the juxtaform superclass and possess a flagellum that is laterally attached to the cell body. Forms d, e and f represent the liberform superclass and do not possess a laterally attached flagellum. Amastigotes (c) exist for both liberform and juxtaform trypanosomatids
Development of Leishmania and Trypanosoma in vertebrates
During a blood meal the infected sand-fly injects metacyclic promastigotes into the vertebrate host [34]. In the vertebrate host, these promastigotes invade macrophages and then differentiate into non-motile amastigotes, which multiply by binary fission [34]. Leishmania replicates within macrophages at various sites depending on the species [10], giving rise to three distinct clinical forms of leishmaniasis; cutaneous leishmaniasis (CL), mucocutaneous leishmaniasis (MCL) and visceral leishmaniasis (VL or Kala Azar) [4]. These clinical forms result from parasite development within the reticuloendothelial system of either the skin (CL), nasopharynx (MCL) or viscera (VL) [35] (Figs. 3, 4).
Some clinical manifestations of leishmaniasis. a A patient with mucocutaneous leishmaniasis (MCL) presenting with facial ulcerative lesions and nasal obstruction. Cropped from original. Citation: Gois et al. Immune response to Leishmania antigens in an AIDS patient with mucocutaneous leishmaniasis as a manifestation of immune reconstitution inflammatory syndrome (IRIS): a case report. BMC Infect Dis. 2015;15(1):38 [154]. b Presentation of MCL with patients suffering from erythematous papules and ulcerations on the lip region. Cropped from original. Citation: Mohammadpour et al. Lip leishmaniasis: a case series with molecular identification and literature review. BMC Infect Dis. 2017;17(1) [155]. c A patient with cutaneous leishmaniasis presenting with crusted nodules over the left cheek (upper panel) and erythematous ulcerated plaques with crusts over the feet (lower panel). Cropped from original. Citation: Al-Dwibe et al. Contact dermatitis-like cutaneous leishmaniasis in a Libyan HIV patient. Parasit Vectors. 2014;7:3 [156]. a-c [157]
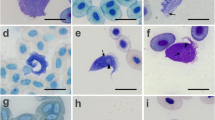
Photomicrographs of stained smears showing Leishmania infections. a May-Grunwald-Giemsa stained preparation from a case of feline leishmaniasis showing macrophages infected with L. infantum amastigotes. Cropped from original. Citation: Pennisi et al. [157]. LeishVet update and recommendations on feline leishmaniosis. Parasit Vectors. 2015;8(1):302. b Haematoxylin and eosin stained histological preparation from a canine deep dermis mucocutaneous lesion showing Leishmania amastigotes and Leishmania-infected fibroblasts (arrowheads). Cropped from original. Citation: Baneth et al. [158]. Mucocutaneous Leishmania tropica infection in a dog from a human cutaneous leishmaniasis focus. Parasit Vectors. 2014;7:5. c Haematoxylin and eosin stained preparation from a canine with cutaneous leishmaniasis showing intracellular Leishmania amastigotes in macrophages (arrows). Cropped from original. Citation: Ordeix et al. [159]. Histological and parasitological distinctive findings in clinically-lesioned and normal-looking skin of dogs with different clinical stages of leishmaniosis. Parasit Vectors. 2017;10:8. d May-Grunwald-Giemsa stained preparation from an aspirate of a mucocutaneous lesion predominately showing intracellular Leishmania amastigotes and few extracellular amastigotes. Cropped from original. Citation: Baneth et al. [158]. a-d Copyright: Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/)
CL is the most common clinical form of the disease, presenting as a myriad of manifestations affecting the skin [8]. It is characterised by the presence of one or more skin lesions with varying morphologies that can result in highly disfiguring scars [8]. Mucocutaneous leishmaniasis is a severely disfiguring form of leishmaniasis with 90% of MCL patients having suffered from a case of CL that had healed 1–5 years prior to the onset of MCL [36]. MCL involves the ulceration of the nasal region, accompanied by symptoms of fever and hepatomegaly [36]. Later progression involves ulceration to the oronasopharyngeal mucosa, resulting in erythema and edema of the affected regions. VL is the most severe manifestation of the disease [8]. VL is a systemic disease, characterised by the incidence of irregular bouts of fever, significant weight loss, splenomegaly and anaemia [8]. If untreated, fatality is almost certain as a result of haemorrhaging or co-infections with bacteria or viruses [4].
Human infective trypanosomes differ significantly in their route of infection, proliferative stages, and preferred sites of infection. Trypanosoma cruzi is transmitted in the faeces of their triatomine vector which defecates on the victim’s skin during a blood meal. The action of biting causes the victim to unknowingly scratch the bite area rubbing metacyclic trypomastigotes of T. cruzi from the faeces into the bite wound or into micro-abrasions caused by scratching [37]. These motile metacyclics invade host cells where they differentiate into amastigotes [4]. These intracellular amastigotes multiply by binary fission, causing tissue damage and host cell apoptosis [38]. Trypanosoma cruzi favours cells of the cardiovascular system, though also affects cells of the nervous and muscular reticuloendothelial systems [3, 39]. Additionally, the oral mode of T. cruzi transmission is emerging as a major route of infection for humans and animals in some endemic regions [40]. Oral transmission occurs via the ingestion of food contaminated with infected triatomine bugs or their faeces. Between 1968 and 2000, 50% of acute cases of Chagas disease recorded from the Brazilian Amazon were attributed to micro-epidemics of the disease caused by oral transmission [40]. Experimental evidence indicates that the infectivity of metacyclic trypomastigotes increases upon contact with gastric juices, a process that is thought to be modulated by certain trypomastigote surface glycoproteins [40, 41]. Alternatively, recent rodent studies suggest that the membranes of the oral mucosa are the most important site of T. cruzi entry into the host via the oral route [42].
Chagas disease has two phases, an acute and chronic phase [5]. In most cases of acute infection, patients are asymptomatic or may experience a wide-range of general symptoms including fever, headaches, heart inflammation, difficulty in breathing, diarrhoea and enlarged lymph glands [16]. Twenty to forty percent of those infected will suffer from severe, irreversible complications during the chronic phase including fatal cardiomyopathy, gastrointestinal and neurological problems [16, 17]. It is important to note that recently, T. cruzi has been divided into six discrete taxonomic units (DTUs) which represent distinct lineages [43, 44]. Recent studies suggest that these DTUs differ in their geographical distribution, transmission and clinical manifestation [44], implying that Trypanosoma cruzi represents a species complex.
Transmission of T. brucei occurs through the bite of the tseste fly, where metacyclic trypomastigotes are injected into the bite wound [4]. Following infection, T. brucei metacyclics transform into blood stream trypomastigotes where they undergo multiplication by binary fission, travelling throughout the blood stream and lymphatic system [45]. Unlike T. cruzi, T. remains extracellular throughout its entire life-cycle.
Human African Trypanosomiasis has two distinct forms of infection, chronic and acute, which are caused by two distinct subspecies of T. brucei [46]. The chronic form (aetiology: Trypanosoma brucei gambiense), is endemic in western and central Africa and is the most common form of Human African Trypanosomiasis (HAT), with humans considered the main reservoir for the disease [12]. The acute infection (aetiology: Trypanosoma brucei rhodesiense), is endemic in eastern and southern Africa and is predominately a zoonotic disease that occasionally affects humans [12]. The clinical manifestations of both acute and chronic HAT are often similar but vary in incubation period and severity. During the early stages of infection the parasites reside in the lymphatic system and blood stream causing fever, general malaise, weakness, lymphadenopathies, endocrine disturbances, musculoskeletal pains, and hepatosplenomegaly [12]. In the acute rhodesiense form, the early stage is often fatal as one tenth of patients do not have access to treatment and die from myocardial involvement as a consequence [46]. In the gambiense form, early stage symptoms are often non-specific including lymphadenopathy and hepatosplenomegaly [46]. The second, later stage of infection occurs following an incubation period of weeks and months in rhodesiense and gambiense infection, respectively. In this stage of the infection, the blood–brain-barrier is compromised, allowing the movement of parasites into the brain where it causes severe neurological manifestation including chronic encephalopathy and finally, coma and death [46].
Development of Phytomonas in plants
The genus Phytomonas includes several plant pathogens that are transmitted by phytophagous insects. The biology and life-cycle of Phytomonas spp. is a poorly understood area of science. Phytomonas has been isolated from 24 different plant families, where they primarily occur as promastigotes and less frequently as choanomastigotes (Fig. 2) [47], while it assumes the form of a slim promastigote in the insect vector [48]. Phytomonas species can infect more than 100 plant species including lactiferous plants, tomato fruits, the coffee tree, coconut and oil palms [49, 50]. Phytomonas spp. have been isolated from a wide-range of tissues including the fruit, flower, seeds and phloem of plants [51]. The genus Phytomonas is restricted to trypanosomatids infecting plants [52], though the ability of Leptomonas seymouri to multiply in plants following experimental infections suggests that this taxonomic criterion should be used with caution and highlights the requirement for molecular evidence when making taxonomic assignments [53].
Development in invertebrates
Monoxenous trypanosomatids
The monoxenous trypanosomatids infect a broad range of insects, including those of the orders Diptera, Hemiptera, Hymenoptera and Siphonaptera [54]. The life-cycle of monoxenous trypanosomatids has only been described for a few species, and current knowledge is based namely on the development of Leptomonas ctenocephali in the flea [55], and Strigomonas oncopelti from the spotted milk-weed bug [56]. Strigomonas oncopelti was originally named Leptomonas oncopelti based on the classical trypanosomatid taxonomic system, though was reassigned to the genus Strigomonas based on phylogenetic analysis [26, 57]. Invertebrate hosts of monoxenous trypanosomatids may become infected via multiple routes including ingestion of cyst-like amastigotes from the faeces of another infected hosts [56], food sharing, predating other infected insect species, or cannibalism [58]. In the posterior portion of the invertebrate (i.e. the insect hindgut), reproduction occurs in one of two ways: binary fission or budding. The former type of reproduction involves nuclear division of promastigotes resulting in daughter cells equal to that of the parent organism. Budding is the mode of reproduction employed by amastigotes, which multiply in the rectum until they are excreted in the faeces [55, 56, 59]. It should be noted that this description of the monoxenous life-cycle represents a generalisation based on knowledge of only a few monoxenous species, and may not apply to all (Fig. 5).
The growth cycle of trypanosomatids within invertebrates. a Replication of Leishmania (Leishmania) species in the sand fly vector occurs at two locations: procyclic promastigotes replicate in the abdominal midgut and leptomonad promastigotes in the thoracic midgut. The replicative procyclic promastigote forms differentiate into elongated nectomonad promastigotes that migrate anteriorly into the thoracic midgut, where further replication in the leptomonad form occurs. Some leptomonad promastigotes attach to the cuticle-lined surface of the midgut and differentiate into haptomonad promastigotes that may act as precursors for differentiation into metacyclic promastigotes, which is the stage infective to the mammalian host. b Leptomonas are ingested in the cyst form and differentiate into the promastigote form. In the crop, the leptomonad form undergoes fission and later in the midgut and pylorus by unequal fission or budding. Cysts are formed via budding in the rectum and are passed out in the faeces as the infective form
Trypanosoma
Trypanosoma species employ one of two methods of development within their invertebrate host, termed Salivaria and Stercoraria [60]. Salivaria as observed in T. brucei and other African trypanosomes, is characterised by development within the frontal portion of the invertebrates’ digestive system and transmitted through the bite of an insect [61]. Stercoraria as observed in Trypanosoma cruzi, is characterised by development of parasites within the posterior region of the invertebrates’ hindgut and transmitted through the excretion of faeces [60]. Trypanosoma evansi, the aetiological agent of the livestock disease Surra, is an example of a trypanosome that is transmitted mechanically [62]. As a result of the loss of mincircles and maxicircles of the kinetoplast DNA [63], T. evansi cannot undergo replication in the tsetse fly and is mechanically transmitted between vertebrates in the mouthparts of blood-feeding flies including the tabanids and stomoxes Trypanosoma equiperdum is unusual amongst trypanosomatids as it is sexually transmitted between horses, causing an illness known as Dourine [62].
Leishmania
Through the detailed study of Leishmania development in sand flies it became increasingly clear that this process was not uniform across the genus. There are several morphological classes of promastigotes that were once used for taxonomic purposes including procyclic, nectomonad, leptomonad, haptomonad and metacyclic promastigotes, discussed later in this paper. A landmark suggestion by Lainson et al. [64] in 1979 established three ‘sections’ discernible by differences in their site of development within the sand fly. These sections greatly aided the classification of Leishmania, and are as follows: (i) Hypopylaria; development in the invertebrate hindgut (restricted to some species that infect only reptiles, i.e. some saurian species), (ii) Peripylaria; development in the hindgut and pylorus, and (iii) Suprapylaria; development anterior to the invertebrate pylorus [65] (Fig. 6). In 1987 species in the Suprapylaria section were assigned to the subgenus Leishmania, i.e. Leishmania (Leishmania), and those in the Peripylaria section were assigned to the subgenus Viannia [19]. This system proved to be an excellent taxonomic criterion, as the three sections were phylogenetically supported [19]. The Hypopylaric Leishmania species are also restricted to lizards; a convenient taxonomic criterion that supports this section, leading to establishment of the genus Sauroleishmania which was eventually demoted to subgeneric status [19, 66]. Debate over which section was the more primitive ensued to decide which section should occupy the root of phylogenetic trees with the Hypopylaric (subgenus Sauroleishmania) and Peripylaric (subgenus Viannia) species representing the prime contenders [66, 67]. Phylogenetic evidence indicates that Hypopylaria does not reflect the most primitive state as saurian species occupy a position closer to the crown of trees than Viannia [68]. In fact, the subgenus Mundinia is the most basal subgenus of Leishmania [28, 68], though knowledge on the development of Mundina in invertebrates is limited.
Diagrammatic representation of the three Leishmania sections proposed by Lainson & Shaw (1979). Figure shows the sections Hypopylaria, Suprapylaria and Peripylaria relative to the relevant structural features of the sand fly including the proboscis (pr), stomodeal valve (sv), cardia (c), thoracic midgut (tm), abdominal midgut (am), malpighian tubules (mt), pylorus (py) and rectum (r). The distribution of Leishmania development within the sand fly vector is shown in black
Light and electron micrographs of Zelonia australiensis. a Transmission electron micrograph showing the gross morphological features of Zelonia australiensis promastigotes including the nucleus (Nu), karysome (Ka), kinetoplast (K), flagella (fl), flagella pocket (fp), glycosomes (gl) and the Golgi body (gb). Subpelicular microtubules (S) give some cell edges a striated appearance, depending on the angle of sectioning. b, c Light micrographs showing promastigotes in a Leishman stained smear. d Light micrograph of a live-cell wet preparation viewed under phase contrast microscopy
In Peripylaria development begins following attachment of parasites to the wall of the sand fly pylorus and ileum (i.e. the hindgut), followed by movement into the midgut and final invasion of the foregut [65, 66]. This section comprises important pathogens of humans and other mammals, including all Viannia parasites and two saurian species; L. (S.) adleri and L. (S.) tarentolae [65, 69]. Members of the Suprapylaria section are limited to growth and differentiation in the midgut and foregut of the sand fly [69], and comprise of the phylogenetically supported subgenus Leishmania and the vast majority of medically significant species [66].
The protein phosphoglycan-rich promastigote secretory gel (PSG) plays a crucial role in Leishmania development within the sand fly by conditioning the gut for differentiation from poorly infective procyclics into highly infective metacyclic promastigotes, a process referred to as metacyclogenesis [70,71,72]. This PSG is secreted by leptomonad promastigotes (Fig. 5) and creates a blockage in the sand fly gut within a week after infection, which is an essential component of Leishmania transmission. The PSG plug forms in the anterior midgut, stomodeal valve and foregut and makes feeding difficult, causing the fly to take prolonged and multiple blood meals from the same host. The plug likely generates backpressure in the gut that causes dislodgement and regurgitation of the plug into the bite wound, along with highly infective metacyclic promastigotes [23, 73]. The formation of a PSG plug is best known in phlebotomine sand flies, though a mass resembling a PSG plug was also observed in Forcipomyia (Lasiohelea) species midges infected with Leishmania macropodum [23, 68].
Problems with classical trypanosomatid systematics
Dixenous and monoxenous clades are paraphyletic
The classical trypanosomatid taxonomic system based on morphology and life-cycles was established in the 1960s by Hoare and Wallace, who redefined kinetoplastid systematics [26, 74, 75]. Early descriptions defined Leptomonas as having a life-cycle containing both promastigote and amastigote stages [76], being parasitic only to invertebrates and of no medical importance [77,78,79]. This system also restricted Leptomonas spp. and other monoxenous genera to a single invertebrate host. This paradigm is no longer accepted as molecular studies have confirmed that many trypanosomatids parasitise multiple insect species [54]. Leptomonas along with other monoxenous genera (Table 1) are often described as “lower trypanosomatids” under the assumption that all trypanosomatids are thought to share a single monoxenous ancestor [80]. While the phrase “lower trypanosomatid” remains in general use, the concept is imperfect as dixenous parasitism has evolved independently multiple times in trypanosomatids, meaning that some dixenous genera (e.g. Trypanosoma) are basal to certain monoxenous species (e.g. Crithidia) [27, 81]. Regardless, it is generally accepted that monoxenous parasitism represents the more primitive, ancestral state as monoxenous species are typically thought to be more numerous and diverse [24, 82]. This does assume however that all trypanosomatid species evolve at the same rate which is thought not to be the case [26, 67]. Regardless, the presumably monoxenous trypanosomatid Paratrypanosoma confusum does support this concept as it invariably occupies the most basal position in molecular phylogenies of trypanosomatids [83].
While monoxenous trypanosomatids are considered non-pathogenic to vertebrates, their restriction to invertebrates is not absolute considering the reports of monoxenous trypanosomatids (e.g. Leptomonas seymouri) exploring the dixenous niche [80, 84,85,86] (discussed below). While these cases are seemingly rare exceptions rather than common events, they highlight that isolation of promastigotes from a vertebrate does not confirm a Leishmania infection which does confound classic taxonomic definitions.
Exploration of the dixenous niche by monoxenous parasites
Of the 19 trypanosomatid genera currently recognised, fourteen are considered monoxenous, parasitising one or more insect hosts [27, 28, 87] (Table 1). Whilst they are considered non-pathogenic to vertebrates, reports of infections with monoxenous species resulting in human illness date back to the 1980s [34, 88] (Table 2). A common trend in many cases was the presence of an immunocompromised state, generally resulting from HIV co-infection [84, 89]. Patients co-infected with HIV and a monoxenous trypanosomatid may present with skin lesions, splenomegaly and other symptoms resembling VL and/or CL [37, 80, 90]. Several cases of infection with Leptomonas spp. have recently come to prominence in HIV negative Indian Kala-azar (VL) patients infected with Leishmania donovani [53, 80, 85, 91]. Whilst the impact of Leptomonas co-infection on the clinical outcome of Kala-azar in these patients remains unexplored, these events confound classic taxonomic definitions and have even led to the incorrect assignment of Leptomonas DNA sequences to L. donovani [85]. The first co-infection involving Leishmania donovani and a monoxenous trypanosomatid was reported in 2010 by Srivastava et al. [91], who employed DNA sequencing to confirm L. donovani infection concurrent with a trypanosomatid designated as Leptomonas sp. BHU [91]. Several similar cases have been reported since, with later studies incriminating Leptomonas seymouri as the opportunistic monoxenous agent [80, 85, 90]. In such cases, Leptomonas may be detected in bone marrow and splenic aspirates, reminiscent of VL [90]. It was theorised that VL induces strong immunosuppression providing the opportunity for L. seymouri to infect these patients [37, 84]. Leptomonas was once considered a primitive sister taxon to Leishmania. In actual fact these genera exhibit relatively little divergence, so it is not unprecedented that L. seymouri might occasionally explore the dixenous niche and possess adaptations that allow it to do so [53, 80]. Another point of contention is that L. seymouri was originally isolated from a strictly phytophagous Dysdercus suturellus “cotton stainer” bug (family Pyrrhocoridae) [53], raising questions surrounding the route of infection in cases of L. donovani - L. seymouri co-infections. Rinsing wounds or sores with water contaminated with L. seymouri is a possibility, and rinsing of syringes in water contaminated with trypanosomatids by intravenous drug abusers is another alternative [84], though drug abuse was not noted in the Indian VL cases. A global study screening the Pyrrhocoridae for trypanosomatids failed to identify L. seymouri parasitising this group [53, 92], suggesting that L. seymouri may possess an alternative hematophagous host.
Infections with ‘monoxenous’ trypanosomatids have also been reported in vertebrates other than humans [86]. For example, an unnamed trypanosomatid was isolated from rats and stray dogs in Egypt in 1989 by Morsey et al. (1988) [93]. A later study by Podlipaev et al. [94] confirmed this parasite as a member of the ‘monoxenous’ genus Herpetomonas, and a very close relative of Herpetomonas ztiplika; a parasite that infects biting midges [94, 95].
Trypanosomatid morphology
Light and electron microscopic studies show that trypanosomatids possess a general ultrastructure that is universally conserved across the family [32]. Trypanosomatids possess an interlaced network of circular DNA known as the kinetoplast DNA (kDNA), associated with the base of a single flagellum that is attached to a slender cell body [32]. Recently, Wheeler et al. [32] conveniently divided trypanosomatids into two distinct morphological superclasses that are supported by molecular phylogeny; the ‘juxtaform’ superclass which includes epimastigote and trypomastigote morphotypes, defined by the presence of a laterally attached flagellum (e.g. Trypanosoma), and the ‘liberform’ superclass which possess a free flagellum (e.g. Leishmania and Leptomonas), which includes the opisthomastigote, choanomastigote and promastigote morphotypes [32] (Fig. 2). The spherical amastigote morphotype of trypanosomatids occurs in both morphological superclasses [32]. Therefore, the presence or absence of amastigotes cannot be used to inform evolutionary relationships. Along with the morphotypes described above, several others have been defined [26].
Liberform trypanosomatids such as Leptomonas, Zelonia (Fig. 7) and Leishmania predominately exist as promastigotes in standard culture, though axenic amastigotes of Leishmania can be induced in vitro [96]. Regardless, these species are highly pleomorphic, shifting between morphotypes depending on their growth phase, host, and host compartment they are occupying [32, 34, 68, 87, 97, 98]. Promastigotes of Leishmania are divided into five morphological categories which include: (i) procyclic promastigotes - the replicating form in the sand fly; (ii) nectomonad promastigotes - the elongated promastigote stage; (iii) haptomonad promastigotes - a stage possessing a disc-like expansion at the flagellar tip; (iv) leptomonad promastigotes - the stage that secretes PSG and is a precursor to; (v) metacyclic promastigotes - the form infective to the vertebrate host (Fig. 5) [70].
Generally, the trypanosomatid morphotypes are distinguished by their cell shape, the relative position of their nucleus to the kinetoplast [33], and flagellum positioning and attachment to the cell body [32, 74, 75]. These and other subtleties once served as taxon-defining characteristics under the classic system, and are now considered to a very minor extent given the inadequacies of this system revealed by genetics [26, 68, 97]. Phylogenetic analyses have culminated in the renaming of several species due to various instances of polyphyly introduced by the classic system, and are now considered mandatory for making accurate taxonomic assignments for new isolates [27, 28, 97, 99, 100].
As new trypanosomatid species are discovered, discernible features that were once taxon defining are becoming less suitable for this purpose. The discovery and description of Novymonous esmeraldas and characterisation of its bacterial endosymbiont provides a recent example [87]. Bacterial endosymbionts were once confined to the Strigomonadinae which occupy a single phylogenetic clade [26]. However, N. esmeraldas is a distantly related trypanosomatid of the subfamily Leishmaniinae, making bacterial endosymbiosis a polyphyletic trait. However, the endosymbionts of the Strigomonadinae are only distantly related to those of Novymonas, suggesting these relationships developed independently in the two trypanosomatid subfamilies [87]. In any case, the main confusion with trypanosomatid systematics lies in the discordance between their visually discernible characteristics and their molecular biology.
Different genera, different rules
Advances in molecular biology and phylogenetics have revealed a lack of taxonomic concordance between gene sequence similarity and species demarcation, most notably within the clinically relevant genera Leishmania and Trypanosoma [7]. For example, small subunit ribosomal RNA (SSU rRNA) sequences of T. cruzi and T. brucei are separated by a genetic distance of approximately 12% while different species within the genus Leishmania possess SSU rRNA sequences separated by a distance of less than 1% in some cases [101]. Similarly, T. cruzi isolates can be separated into several discrete typing units [102, 103], which could easily constitute different species if they were held to the same species demarcation criteria as the genus Leishmania. This problem has been raised by previous investigators, who offer sensible solutions to this issue, including delineation of new species based on a 90% sequence similarity threshold [27].
Hybridization between species
Based on current understanding Leishmania possesses a sexual or parasexual cycle that allows recombination between distinct lineages, and in some cases different species [80]. Instances of hybridisation have been known from studies of trypanosomatid field isolates for nearly two decades [104]. A study involving isoenzyme analysis and molecular karyotyping of two Leishmania strains isolated from wild animals in Saudi Arabia identified a suspected hybrid isolate distinct from other Leishmania species, possessing characteristics of both Leishmania major and Leishmania arabica [105]. Banuls et al. [106] identified suspected Leishmania hybrids in stocks isolated from humans in Ecuador, representing potential crosses between L. braziliensis and L. panamensis/guyanensis. More recently, whole genome sequencing was used to confirm hybridisation in 11 unique isolates of Leishmania infantum from sand flies and one CL patient [107]. Additionally, crosses between L. major and L. infantum have been achieved under experimental conditions [108]. The limited genetic divergence between Leishmania species (discussed above), in conjunction with reports of hybridization, indicate that species delineation in this genus could be relaxed. However, echoing the points made by previous investigators [27], collapsing the many Leishmania species into a few would only generate confusion, particularly for clinicians. The grouping of Leishmania species into phylogenetically supported subfamilies as per Espinosa et al. [28] seems an elegant solution as hybridization seems to only occur within subfamilies and not between them, though this requires further investigation.
Exploration of alternative vectors
Cases of locally acquired Australian cutaneous leishmaniasis were first reported in 2004, in captive red kangaroos (Macropus rufus) from a wildlife park near Darwin, in Australia’s Northern Territory [20, 109]. In 2009, the same species was isolated from three additional macropod species, Macropus robustus woodwardi (northern wallaroo), Macropus bernardus (black wallaroo) and Macropus agilis (agile wallaby) [22]. This was the first report confirming Australia’s endemicity for leishmaniasis, albeit a form restricted to native animals, with no evidence for human infection. Of the 18 known Australian phlebotomine species, little is known about their biological capacity as vectors of leishmaniasis and Leishmania has never been detected in these species [23, 110]. Australian phlebotamines are thought to feed solely on small mammals, birds and reptiles which led to the speculation that this species (recently dubbed Leishmania macropodum [68]), might be transmitted via an alternative vector [22]. Later investigations identified a day feeding midge, Forcipomyia (Lasiohelea) (Diptera: Ceratopogonidae), as the likely vector [23]. Leishmania macropodum was prevalent in ~15% of Forcipomyia (L.) midges tested and similar patterns of promastigote migration to the midgut were observed between Forcipomyia and Leishmania, when compared to those observed in the Leishmania-phlebotomine interaction [23]. In addition to Forcipomyia, other insects may be capable of supporting Leishmania replication (at least temporarily) based on evidence from experimental infections and molecular evidence from naturally infected insects. The biting midge Culicoides sonorensis (Diptera: Ceratopogonidae) is capable of developing late stage infections with Leishmania enriettii [111]. Culicoides nubeculosus supported L. infantum infection for up to 7 days post blood meal [72]. Leishmania infantum DNA has also been detected in naturally infected Culicoides spp. collected in Tunisia [112], while DNA from Leishmania amazonensis and Leishmania braziliensis was detected in naturally infected Culicoides spp. from Brazil [113]. These studies call into question the exclusivity of the Leishmania-phlebotomine interaction, particularly in the case of the Australian species which naturally infects Forcipomyia and reportedly produces a PSG plug in this insect [23].
The phylogenetic solution and its caveats
Phylogenetic evidence has confirmed on multiple occasions that naming trypanosomatid taxa based on classic systems often gives rise to polyphyly [99, 114]. This is due to divergence in DNA sequences that give rise to few changes in morphology, host range and other biological characteristics [7, 97]. The absence of adequate boundaries relating to morphological and host-based criteria highlight the requirement for a phylogenetic solution to trypanosomatid taxonomy [115]. Estimating rates of evolutionary divergence is attributed to Zuckerkandl and Pauling’s concept of the molecular clock which suggests that differences between amino acid sequences are relatively proportional to evolutionary events of divergence [116,117,118]. This approach assumes that rates of genetic change are constant amongst species of common descent, allowing estimates of rates to be extrapolated across phylogenetic trees [119]. Subsequent studies suggested that if these clocks could infer evolutionary timescales, the differences between sequences must be exclusive to sites of neutrality to equal the rate of mutation, supporting Kimura’s theory of neutral molecular evolution [120, 121].
Slow evolving (SE) genes are characterised by a slower divergence rate, undergoing fewer nucleotide substitutions over time. Sequences of SE genes are most suitable for investigating evolutionary relationships over larger time-scales [122]. Phylogenetic studies using only the small subunit ribosomal RNA (SSU rRNA) genes may not provide reliable phylogenetic inference for species within the same taxonomic family for example, as the rRNA genes evolve very slowly [123, 124]. Instead, a middle ground must be reached for resolving relationships between closely related organisms. Several housekeeping genes encoding proteins involved in basic cellular functions have provided the most useful information on relationships between the various Leishmania species [125]. Gene sequences of the RNA polymerase II largest subunit (RPOIILS) [97, 123], DNA polymerase α catalytic polypeptide (POLA) [123], glyceraldehyde-3-phosphate dehydrogenase (GADPH) [18, 97], heat-shock protein 20 [125], and heat-shock protein 70 [126], are preferred targets for analysing phylogenetic relationships between Leishmania species and their close relatives.
Despite the power of phylogenetic inference there are some important caveats to be considered. For example, the structure of phylogenetic trees can change markedly depending on the locus used, affecting the way relationships between taxa are interpreted. Using concatenated sequences from multiple phylogenetically informative loci has been suggested as a means to counteract this problem and improve the robustness of phylogenetic trees [27]. Similarly, the structure of phylogenetic trees can change markedly depending on the outgroup selected, or if certain taxa are included or excluded from the analysis [27]. The robustness of phylogenetic predictions is also dependent on the number of appropriate taxa included in the analysis. An important example involves the early debate surrounding the most basal Leishmania subgenus (touched upon previously herein) [19, 67, 68, 127,128,129,130]. This was considered pertinent to selection of the most appropriate outgroup, and would have implications relating to Leishmania biogeography and even its first vertebrate hosts. If the Sauroleishmania were the earliest branching group, this would implicate reptiles as the original vertebrate host of an ancestral Leishmania parasite. If Viannia were most basal, Leishmania probably evolved in the Neotropics [128]. With the addition of several other taxa, including some novel monoxenous genera that represent appropriate outgroups; phylogenetic evidence indicates that Mundinia is the most basal Leishmania subgenus [28, 68].
Discrepancies also arise when using single (or very few) trypanosomatid isolates to define new genera and establish taxonomic assignments. Zelonia costaricensis represents one such example. Originally assigned to the genus Leptomonas [131], the inclusion of several novel trypanosomatid isolates in phylogenies confirmed that Zelonia is distinct from the Leptomonas, Lotmaria and Crithidia clade, but is immediately basal to the Leishmania, Endotrypanum and Porcisia clade, warranting establishment of a new genus [28, 68]. A solution exemplified by Espinosa et al. [28] is to obtain sequence data from multiple sister isolates of a new candidate taxon before making new assignments. This will ensure that trypanosomatid taxa remain monophyletic by providing robust phylogenetic support.
Two recent studies used phylogenetics to approximately date the origin of the first ancestral Leishmania parasites with intriguing results [68, 132]. However, this type of analysis is complicated by the fact that closely related taxa may evolve at different rates due to environmental pressures, including those exerted by the host immune response. These pressures are of course markedly different for monoxenous and dixenous taxa. The accuracy of these analyses also relies on selection of a precise calibration point; that is, an accurately dated geological event (or other) that is known to have triggered a speciation event in closely related taxa. As the accuracy of these predictions are difficult to test, vicariance events dated using phylogenetic inference should be considered approximations at best.
Conclusions
Important developments have been made in the field of trypanosomatid taxonomy in recent years, facilitated by the identification of novel trypanosomatid species in addition to advances in molecular biology and phylogenetics. Phylogenetic inference has identified flaws in the classical system of trypanosomatid taxonomy which relied predominantly on parasite morphology and host preferences. Despite significant sequence divergence, trypanosomatids have experienced limited morphological change throughout their evolution such that the subtle morphological variations that exist do not reflect molecular phylogeny. Phylogenetics has corrected some of these issues by supporting the assignment of some trypanosomatids to different genera and establishment of new genera, ensuring that taxa remain monophyletic [27]. Despite these advances, trypanosomatid systematics still remains imperfect. There remains a lack of consensus for delineating species based on molecular divergence, and the limits differ markedly for Trypanosoma and Leishmania. While not recommended due to the confusion it would cause, molecular evidence probably supports the collapsing of some Leishmania species into one, or splitting Trypanosoma into different species or even different genera depending on the limits of molecular divergence applied. Additionally, some trypanosomatid genera (i.e. Leptomonas) remain paraphyletic [68, 133]. Transmission of L. macropodum by a day feeding midge rather than a phlebotomine sand fly and the detection of clinically important Leishmania species in hematophagous insects such as Culicoides spp., questions the exclusivity of the Leishmania-sand fly relationship. Reports of monoxenous trypanosomatid infections in vertebrates and the grouping of presumably monoxenous trypanosomatids (e.g. Zelonia and Novymonas) in dixenous clades occupied by Leishmania and Endotrypanum, also call into question the suitability of host relationships as a taxonomic criterion. Long accepted dogmas such as the ‘one-insect-one-parasite’ rule have been overturned, supported by molecular evidence. These examples highlight the importance of employing molecular biology and phylogenetics when making taxonomic assignments relating to trypanosomatids. The continued isolation and characterisation of novel trypanosomatids will ensure this field continues to develop, by allowing resolution of evolutionary relationships between members of this group with increasing accuracy. To this end, generation of high-quality genome sequences for additional species would be of huge benefit, allowing relationships to be phylogenetically scrutinised across numerous loci. The genomes of trypanosomatids that encroach on the monoxenous-dixenous boundary (e.g. Zelonia and Novymonas) may prove instrumental to understanding the path to dixenous parasitism by providing new knowledge on the innovations that evolved to allow this transition [58].
Abbreviations
- CL:
-
Cutaneous leishmaniasis
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- Hsp20:
-
Heat-shock protein 20
- Hsp70:
-
Heat-shock protein 70
- MCL:
-
Mucocutaneous leishmaniasis
- POLA:
-
DNA polymerase α catalytic polypeptide
- PSG:
-
Promastigote secretory gel
- RPOIILS:
-
RNA polymerase II largest subunit
- SE:
-
Slow evolving
- SSU rRNA:
-
Small subunit ribosomal RNA
- VL:
-
Visceral leishmaniasis
References
Overath P, Haag J, Lischke A, O’HUigin C. The surface structure of trypanosomes in relation to their molecular phylogeny. Int J Parasitol. 2001;31(5–6):468–71.
Bouteille B, Oukem O, Bisser S, Dumas M. Treatment perspectives for human African trypanosomiasis. Fundam Clin Pharmacol. 2003;17(2):171–81.
Hemmige V, Tanowitz H, Sethi A. Trypanosoma cruzi infection: a review with emphasis on cutaneous manifestations. Int J Dermatol. 2012;51(5):501–8.
Nussbaum K, Honek J, Cadmus C, Efferth T. Trypanosomatid parasites causing neglected diseases. Curr Med Chem. 2010;17(15):1594–617.
Rassi A, Rassi A, de Rezende JM. American trypanosomiasis (Chagas disease). Infect Dis Clin North Am. 2012;26(2):275–91.
Stewardson AJ, Leder K, Torresi J, Johnson DF. Two cases of Old World cutaneous leishmaniasis in Australian travelers visiting Morocco. J Travel Med. 2010;17(4):278–80.
Vickerman K. The evolutionary expansion of the trypanosomatid flagellates. Int J Parasitol. 1994;24(8):1317–31.
WHO. Research priorities for Chagas disease, human African trypanosomiasis and leishmaniasis. WHO Tech Rep Ser. 2012;975(5):11–9.
Baldwin T, Henri S, Curtis J, O’Keeffe M, Vremec D, Shortman K, et al. Dendritic cell populations in Leishmania major-infected skin and draining lymph nodes. Infect Immun. 2004;72(4):1991–2001.
Baldwin TM, Elso C, Curtis J, Buckingham L, Handman E. The site of Leishmania major infection determines disease severity and immune responses. Infect Immun. 2003;71(12):6830–4.
Morrison LJ. Parasite-driven pathogenesis in Trypanosoma brucei infections. Parasite Immunol. 2011;33(8):448–55.
Franco JR, Simarro PP, Diarra A, Jannin JG. Epidemiology of human African trypanosomiasis. Clin Epigenetics. 2014;6:257–75.
Stevens JR, Noyes H, Dover GA, Gibson WC. The ancient and divergent origins of the human pathogenic trypanosomes, Trypanosoma brucei and T. cruzi. Parasitology. 1999;118:107–16.
Fermino BR, Paiva F, Soares P, Tavares LE, Viola LB, Ferreira RC, et al. Field and experimental evidence of a new caiman trypanosome species closely phylogenetically related to fish trypanosomes and transmitted by leeches. Int J Parasitol Parasites Wildl. 2015;4(3):368–78.
Human African Trypanosomiasis [http://www.who.int/trypanosomiasis_african/country/en/]. Accssed 25 May 2017.
Chagas disease (American trypanosomiasis) [http://www.who.int/chagas/]. Accssed 25 May 2017.
Jackson Y, Pinto A, Pett S. Chagas disease in Australia and New Zealand: risks and needs for public health interventions. Trop Med Int Health. 2014;19(2):212–8.
Marcili A, Speranca MA, da Costa AP, Madeira MD, Soares HS, Sanches C, et al. Phylogenetic relationships of Leishmania species based on trypanosomatid barcode (SSU rDNA) and gGAPDH genes: taxonomic revision of Leishmania (L.) infantum chagasi in South America. Infect Genet Evol. 2014;25:44–51.
Akhoundi M, Kuhls K, Cannet A, Votýpka J, Marty P, Delaunay P, et al. A historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl Trop Dis. 2016;10(3):e0004349.
Rose K, Curtis J, Baldwin T, Mathis A, Kumar B, Sakthianandeswaren A, et al. Cutaneous leishmaniasis in red kangaroos: isolation and characterisation of the causative organisms. Int J Parasitol. 2004;34(6):655–64.
Best MP, Ash A, Bergfeld J, Barrett J. The diagnosis and management of a case of leishmaniosis in a dog imported to Australia. Vet Parasitol. 2014;202(3–4):292–5.
Dougall A, Shilton C, Choy JL, Alexander B, Walton S. New reports of Australian cutaneous leishmaniasis in Northern Australian macropods. Epidemiol Infect. 2009;137(10):1516–20.
Dougall AM, Alexander B, Holt DC, Harris T, Sultan AH, Bates PA, et al. Evidence incriminating midges (Diptera: Ceratopogonidae) as potential vectors of Leishmania in Australia. Int J Parasitol. 2011;41(5):571–9.
Podlipaev S. The more insect trypanosomatids under study-the more diverse Trypanosomatidae appears. Int J Parasitol. 2001;31(5–6):648–52.
Yurchenko V, Votýpka J, Tesařová M, Klepetková H, Kraeva N, Jirků M, et al. Ultrastructure and molecular phylogeny of four new species of monoxenous trypanosomatids from flies (Diptera: Brachycera) with redefinition of the genus Wallaceina. Folia Parasitol. 2014;61(2):97–112.
Maslov DA, Votýpka J, Yurchenko V, Lukeš J. Diversity and phylogeny of insect trypanosomatids: all that is hidden shall be revealed. Trends Parasitol. 2013;29(1):43–52.
Votýpka J, d’Avila-Levy CM, Grellier P, Maslov DA, Lukeš J, Yurchenko V. New approaches to systematics of Trypanosomatidae: criteria for taxonomic (re)description. Trends Parasitol. 2015;31(10):460–9.
Espinosa OA, Serrano MG, Camargo EP, Teixeira MM, Shaw JJ. An appraisal of the taxonomy and nomenclature of trypanosomatids presently classified as Leishmania and Endotrypanum. Parasitology. 2016;15:1–13. doi:10.1017/S0031182016002092.
Momen H. Some current problems in the systematics of trypanosomatids. Int J Parasitol. 2001;31(5–6):640–2.
Simpson AGB, Stevens JR, Lukeš J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22(4):168c74.
Stevens JR, Noyes HA, Schofield CJ, Gibson W. The molecular evolution of Trypanosomatidae. Adv Parasitol. 2001;48:1–56.
Wheeler RJ, Gluenz E, Gull K. The limits on trypanosomatid morphological diversity. PLoS One. 2013;8(11):18.
Yurchenko VY, Lukeš J, Tesařová M, Jirků M, Maslov DA. Morphological discordance of the new trypanosomatid species phylogenetically associated with the genus Crithidia. Protist. 2008;159(1):99–114.
Grimaldi G, Schottelius J. Leishmaniases - their relationships to monoxenous and dixenous trypanosomatids. Med Microbiol Immunol. 2001;190(1–2):3–8.
Torres-Guerrero E, Quintanilla-Cedillo M, Ruiz-Esmenjaud J, Arenas R. Leishmaniasis: a review. F1000Research. 2017;6(750):1-15.
David CV, Craft N. Cutaneous and mucocutaneous leishmaniasis. Dermatol Ther. 2009;22(6):491–502.
Barratt JL, Harkness J, Marriott D, Ellis JT, Stark D. Importance of nonenteric protozoan infections in immunocompromised people. Clin Microbiol Rev. 2010;23(4):795–836.
Bienvenu AL, Gonzalez-Rey E, Picot S. Apoptosis induced by parasitic diseases. Parasit Vectors. 2010;3:106.
Caradonna KL, Engel JC, Jacobi D, Lee CH, Burleigh BA. Host metabolism regulates intracellular growth of Trypanosoma cruzi. Cell Host Microbe. 2013;13(1):108–17.
Yoshida N. Trypanosoma cruzi infection by oral route How the interplay between parasite and host components modulates infectivity. Parasitol Int. 2008;57(2):105–9.
Cortez M, Silva MR, Neira I, Ferreira D, Sasso GRS, Luquetti AO, et al. Trypanosoma cruzi surface molecule gp90 downregulates invasion of gastric mucosal epithelium in orally infected mice. Microbes Infect. 2006;8(1):36–44.
Silva-Dos-Santos D, Barreto-de-Albuquerque J, Guerra B, Moreira OC, Berbert LR, Ramos MT, et al. Unraveling Chagas disease transmission through the oral route: gateways to Trypanosoma cruzi infection and target tissues. PLoS Negl Trop Dis. 2017;11(4):e0005507.
Zafra G, Mantilla JC, Valadares HM, Macedo AM, Gonzalez CI. Evidence of Trypanosoma cruzi II infection in Colombian chagasic patients. Parasitol Res. 2008;103(3):731–4.
Villa LM, Guhl F, Zabala D, Ramirez JD, Urrea DA, Hernandez DC, et al. The identification of two Trypanosoma cruzi I genotypes from domestic and sylvatic transmission cycles in Colombia based on a single polymerase chain reaction amplification of the spliced-leader intergenic region. Mem Inst Oswaldo Cruz. 2013;108(7):932–5.
Matthews KR. The developmental cell biology of Trypanosoma brucei. J Cell Sci. 2005;118(2):283–90.
Stich A, Abel PM, Krishna S. Human African trypanosomiasis. BMJ. 2002;325(7357):203–6.
Camargo EP. Phytomonas and other trypanosomatid parasites of plants and fruit. Adv Parasitol. 1999;42:29–112.
Seward EA, Votýpka J, Kment P, Lukeš J, Kelly S. Description of Phytomonas oxycareni n. sp. from the salivary glands of Oxycarenus lavaterae. Protist. 2017;168(1):71–9.
Desouza W, Attias M. Cell biology of Phytomonas, trypanosomatids parasites of plants. Mem Inst Oswaldo Cruz. 1991;86(3):275–84.
Porcel BM, Denoeud F, Opperdoes F, Noel B, Madoui MA, Hammarton TC, et al. The streamlined genome of Phytomonas spp. relative to human pathogenic kinetoplastids reveals a parasite tailored for plants. PLoS Genet. 2014;10(2):24.
Jaskowska E, Butler C, Preston G, Kelly S. Phytomonas: trypanosomatids adapted to plant environments. PLoS Pathog. 2015;11(1):17.
Zanetti A, Ferreira RC, Serrano MG, Takata CS, Campaner M, Attias M, et al. Phytomonas (Euglenozoa: Trypanosomatidae): phylogenetic analyses support infrageneric lineages and a new species transmitted to Solanaceae fruits by a pentatomid hemipteran. Eur J Protistol. 2016;56:232–249.
Kraeva N, Butenko A, Hlavacova J, Kostygov A, Myskova J, Grybchuk D, et al. Leptomonas seymouri: adaptations to the dixenous life cycle analyzed by genome sequencing, transcriptome profiling and co-infection with Leishmania donovani. PLoS Pathog. 2015;11(8):23.
Kozminsky E, Kraeva N, Ishemgulova A, Dobáková E, Lukeš J, Kment P, et al. Host-specificity of monoxenous trypanosomatids: statistical analysis of the distribution and transmission patterns of the parasites from neotropical Heteroptera. Protist. 2015;166(5):551–68.
Gibbs AJ. Observations on Leptomonas ctenocephali (Fantham, 1912). Trans R Soc Trop Med Hyg. 1947;40(4):495-500.
Olsen OW. Animal parasites: their life cycles and ecology. 1986. New York Courier Corporation.
Teixeira MM, Borghesan TC, Ferreira RC, Santos MA, Takata CS, Campaner M, et al. Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of proteobacterial symbionts. Protist. 2011;162(3):503–24.
Flegontov P, Butenko A, Firsov S, Kraeva N, Elias M, Field MC, et al. Genome of Leptomonas pyrrhocoris: a high-quality reference for monoxenous trypanosomatids and new insights into evolution of Leishmania. Sci Rep. 2016;6:23704.
Romeiro A, Sole-Cava A, Sousa MA, De Souza W, Attias M. Ultrastructural and biochemical characterization of promastigote and cystic forms of Leptomonas wallacei n. sp. isolated from the intestine of its natural host Oncopeltus fasciatus (Hemiptera: Lygaeidae). J Eukaryot Microbiol. 2000;47(3):208–20.
Stevens J, Gibson W. The evolution of Salivarian trypanosomes. Mem Inst Oswaldo Cruz. 1999;94(2):225–6.
Haag J, O’HUigin C, Overath P. The molecular phylogeny of trypanosomes: evidence for an early divergence of the Salivaria. Mol Biochem Parasitol. 1998;91(1):37–49.
Desquesnes M, Holzmuller P, Lai DH, Dargantes A, Lun ZR, Jittaplapong S. Trypanosoma evansi and Surra: a review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. Biomed Res Int. 2013;2013:1–22.
Lai DH, Hashimi H, Lun ZR, Ayala FJ, Lukeš J. Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc Natl Acad Sci U S A. 2008;105(6):1999–2004.
Lainson R, Ready PD, Shaw JJ. Leishmania in phlebotomid sandflies. VII. On the taxonomic status of Leishmania peruviana, causative agent of Peruvian ‘uta’, as indicated by its development in the sandfly, Lutzomyia longipalpis. Proc R Soc Lond B Biol Sci. 1979;206(1164):307–18.
Peters W, Killick-Kendrick R. The leishmaniases in biology and medicine vol. 1. London: Academic; 1987.
Eldridge BF, Edman J. Medical Entomology: a textbook on public health and veterinary problems caused by arthropods, vol. 1. California: Springer Science & Business Media; 2000.
Kerr SF. Palaearctic origin of Leishmania. Mem Inst Oswaldo Cruz. 2000;95(1):75–80.
Barratt J, Kaufer A, Peters B, Craig D, Lawrence A, Roberts T, et al. Isolation of novel trypanosomatid, Zelonia australiensis sp. nov. (Kinetoplastida: Trypanosomatidae) provides support for a Gondwanan origin of dixenous parasitism in the Leishmaniinae. PLoS Negl Trop Dis. 2017;11(1):e0005215.
Kreier J. Parasitic Protozoa, vol. 10. Ohio: Academic; 1995.
Rogers ME, Chance ML, Bates PA. The role of promastigote secretory gel in the origin and transmission of the infective stage of Leishmania mexicana by the sandfly Lutzomyia longipalpis. Parasit. 2002;124(Pt 5):495–507.
Muskus CE, Marin Villa M. [Metacyclogenesis: a basic process in the biology of Leishmania]. Biomedica: rev del Inst Nac de Salud. 2002;22(2):167–177
Seblova V, Sadlova J, Carpenter S, Volf P. Development of Leishmania parasites in Culicoides nubeculosus (Diptera: Ceratopogonidae) and implications for screening vector competence. J Med Entomol. 2012;49(5):967–70.
Bates PA. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int J Parasitol. 2007;37(10–3):1097–106.
Hoare CA, Wallace FG. Developmental stages of trypanosomatid flagellates: a new terminology. Nature. 1966;212(5068):1385–6.
Wallace FG. The trypanosomatid parasites of insects and arachnids. Exp Parasitol. 1966;18(1):124–93.
Lee J, Leedale G, Bradbury P. Illustrated Guide to the Protozoa. Lawrence: Wiley-Blackwell; 2000.
Roberts L, Janovy J. Foundations of parasitology. 5th ed. London: Wm. C. Brown; 1996.
Molyneux D, Ashford R. The biology of Trypanosoma and Leishmania, parasites of man and domestic animals. London: Taylor and Francis; 1983.
Chauhan IS, Kaur J, Krishna S, Ghosh A, Singh P, Siddiqi MI, et al. Evolutionary comparison of prenylation pathway in kinetoplastid Leishmania and its sister Leptomonas. BMC Evol Biol. 2015;15:19.
Singh N, Chikara S, Sundar S. SOLiD sequencing of genomes of clinical isolates of Leishmania donovani from India confirm Leptomonas co-infection and raise some key questions. PLoS One. 2013;8(2):10.
Frolov AO, Malysheva MN, Yurchenko V, Kostygov AY. Back to monoxeny: Phytomonas nordicus descended from dixenous plant parasites. Eur J Protistol. 2016;52:1–10.
Santos ALS, Branquinha MH, D’Avila-Levy CM. The ubiquitous gp63-like metalloprotease from lower trypanosomatids: in the search for a function. An Acad Bras Cienc. 2006;78(4):687–714.
Flegontov P, Votýpka J, Skalický T, Logacheva MD, Penin AA, Tanifuji G, et al. Paratrypanosoma is a novel early-branching trypanosomatid. Curr Biol. 2013;23(18):1787–93.
Chicharro C, Alvar J. Lower trypanosomatids in HIV/AIDS patients. Ann Trop Med Parasitol. 2003;97:75–8.
Ghosh S, Banerjee P, Sarkar A, Datta S, Chatterjee M. Coinfection of Leptomonas seymouri and Leishmania donovani in Indian Leishmaniasis. J Clin Microbiol. 2012;50(8):2774–8.
Lukeš J, Skalický T, Týč J, Votýpka J, Yurchenko V. Evolution of parasitism in kinetoplastid flagellates. Mol Biochem Parasitol. 2014;195(2):115–22.
Kostygov AY, Dobáková E, Grybchuk-Ieremenko A, Váhala D, Maslov DA, Votýpka J, et al. Novel trypanosomatid-bacterium association: evolution of endosymbiosis in action. MBio. 2016;7(2):e01985-15.
McGhee RB, Cosgrove WB. Biology and physiology of the lower trypanosomatidae. Microbiol Rev. 1980;44(1):140–73.
Pacheco RS, Marzochi MCA, Pires MQ, Brito CMM, Madeira MD, Barbosa-Santos EGO. Parasite genotypically related to a monoxenous trypanosomatid of dog’s flea causing opportunistic infection in an HIV positive patient. Mem Inst Oswaldo Cruz. 1998;93(4):531–7.
Selvapandiyan A, Ahuja K, Puri N, Krishnan A. Implications of co-infection of Leptomonas in visceral leishmaniasis in India. Parasitology. 2015;142(14):1657–62.
Srivastava P, Prajapati VK, Vanaerschot M, Van der Auwera G, Dujardin JC, Sundar S. Detection of Leptomonas sp. parasites in clinical isolates of Kala-azar patients from India. Infect Genet Evol. 2010;10(7):1145–50.
Votýpka J, Klepetková H, Yurchenko VY, Horák A, Lukeš J, Maslov DA. Cosmopolitan distribution of a trypanosomatid Leptomonas pyrrhocoris. Protist. 2012;163(4):616–31.
Morsy TA, Schnur LF, Feinsod FM, Michael SA, Saah A, Salama MM, et al. The discovery and preliminary characterization of a novel trypanosomatid parasite from Rattus norvegicus and stray dogs from Alexandria, Egypt. Ann Trop Med Parasitol. 1988;82(5):437–44.
Podlipaev SA, Sturm NR, Fiala I, Fernanades O, Westenberger SJ, Dollet M, et al. Diversity of insect trypanosomatids assessed from the spliced leader RNA and 5S rRNA genes and intergenic regions. J Eukaryot Microbiol. 2004;51(3):283–90.
Borghesan TC, Ferreira RC, Takata CS, Campaner M, Borda CC, Paiva F, et al. Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist. 2013;164(1):129–52.
Nasereddin A, Schweynoch C, Schonian G, Jaffe CL. Characterization of Leishmania (Leishmania) tropica axenic amastigotes. Acta Trop. 2010;113(1):72–9.
Yurchenko V, Lukeš J, Xu X, Maslov DA. An integrated morphological and molecular approach to a new species description in the trypanosomatidae: The case of Leptomonas podlipaevi n. sp., a parasite of Boisea rubrolineata (Hemiptera: Rhopalidae). J Eukaryot Microbiol. 2006;53(2):103–11.
Walters LL. Leishmania differentiation in natural and unnatural sand fly hosts. J Eukaryot Microbiol. 1993;40(2):196–206.
Merzlyak E, Yurchenko V, Kolesnikov AA, Alexandrov K, Podlipaev SA, Maslov DA. Diversity and phylogeny of insect trypanosomatids based on small subunit rRNA genes: polyphyly of Leptomonas and Blastocrithidia. J Eukaryot Microbiol. 2001;48(2):161–9.
Jirků M, Yurchenko VY, Lukeš J, Maslov DA. New species of insect trypanosomatids from Costa Rica and the proposal for a new subfamily within the Trypanosomatidae. J Eukaryot Microbiol. 2012;59(6):537–47.
Berchtold M, Philippe H, Breunig A, Brugerolle G, Konig H. The phylogenetic position of Dimastigella trypaniformis within the parasitic kinetoplastids. Parasitol Res. 1994;80(8):672–9.
Breniere SF, Waleckx E, Barnabe C. Over six thousand Trypanosoma cruzi strains classified into discrete typing units (DTUs): Attempt at an inventory. PLoS Negl Trop Dis. 2016;10(8):e0004792.
Barnabe C, Mobarec HI, Jurado MR, Cortez JA, Breniere SF. Reconsideration of the seven discrete typing units within the species Trypanosoma cruzi, a new proposal of three reliable mitochondrial clades. Infect Genet Evol. 2016;39:176–86.
Tibayrenc M, Ayala FJ. Evolutionary genetics of Trypanosoma and Leishmania. Microbes Infect. 1999;1(6):465–72.
Kelly JM, Law JM, Chapman CJ, Vaneys G, Evans DA. Evidence of genetic recombination in Leishmania. Mol Biochem Parasitol. 1991;46(2):253–63.
Banuls AL, Guerrini F, LePont F, Barrera C, Espinel I, Guderian R, et al. Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J Eukaryot Microbiol. 1997;44(5):408–11.
Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, Svobodová M, et al. Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet. 2014;10(1):14.
Romano A, Inbar E, Debrabant A, Charmoy M, Lawyer P, Ribeiro-Gomes F, et al. Cross-species genetic exchange between visceral and cutaneous strains of Leishmania in the sand fly vector. Proc Natl Acad Sci USA. 2014;111(47):16808–13.
Seblova V, Sadlova J, Carpenter S, Volf P. Speculations on biting midges and other bloodsucking arthropods as alternative vectors of Leishmania. Parasit Vectors. 2014;7:2.
Stein J, Dyce AL. Field observations on the phlebotomine sand fly Australophlebotomus mackerrasi Lewis and Dyce feeding on the Gidgee skink Egernia stokesii Gray. Parasitol Res. 2002;88(3):278–9.
Seblova V, Sadlova J, Vojtkova B, Votýpka J, Carpenter S, Bates PA, et al. The biting midge Culicoides sonorensis (Diptera: Ceratopogonidae) is capable of developing late stage infections of Leishmania enriettii. PLoS Negl Trop Dis. 2015;9(9):15.
Slama D, Haouas N, Remadi L, Mezhoud H, Babba H, Chaker E. First detection of Leishmania infantum (Kinetoplastida: Trypanosomatidae) in Culicoides spp. (Diptera: Ceratopogonidae). Parasit Vectors. 2014;7:51.
Rebelo JM, Rodrigues BL, Bandeira MD, Moraes JL, Fonteles RS, Pereira SR. Detection of Leishmania amazonensis and Leishmania braziliensis in Culicoides (Diptera, Ceratopogonidae) in an endemic area of cutaneous leishmaniasis in the Brazilian Amazonia. J Vector Ecol. 2016;41(2):303–8.
Olsen GJ, Woese CR. Ribosomal RNA: a key to phylogeny. Fas J. 1993;7(1):113–23.
Schönian G, Cupolillo E, Mauricio I. Molecular evolution and phylogeny of Leishmania. In: Ponte-Sucre A, Diaz E, Padrón-Nieves M, editors. Drug resistance in Leishmania parasites. Vienna: Springer; 2013. p. 15–44.
Zuckerkandl E, Pauling L. Molecular disease, evolution, and genic heterogeneity Horiz. Biochem. 1962; 189–225
Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. Evolv Gen Prot. 1965; 97–166
Wilke T, Schultheiss R, Albrecht C. As time goes by: a simple fool’s guide to molecular clock approaches in invertebrates. Amer Malaco Bull. 2009;27(1–2):25–45.
Yi SV. Understanding neutral genomic molecular clocks. Evol Bioinforma. 2007;34(3–4):144–51.
Nei M, Suzuki Y, Nozawa M. The neutral theory of molecular evolution in the genomic era. In: Chakravarti A, Green E, editors. Annual review of genomics and human genetics, vol. 11. Palo Alto: Annual Reviews; 2010. p. 265–89.
Takahata N. Molecular clock: an anti-neo-Darwinian legacy. Genetics. 2007;176(1):1–6.
Hamilton PB, Stevens JR, Gaunt MW, Gidley J, Gibson WC. Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int J Parasitol. 2004;34(12):1393–404.
Croan DG, Morrison DA, Ellis JT. Evolution of the genus Leishmania revealed by comparison of DNA and RNA polymerase gene sequences. Mol Biochem Parasitol. 1997;89(2):149–59.
Deschamps P, Lara E, Marande W, Lopez-Garcia P, Ekelund F, Moreira D. Phylogenomic analysis of kinetoplastids supports that trypanosomatids arose from within bodonids. Mol Biol Evol. 2011;28(1):53–8.
Fraga J, Montalvo AM, Van der Auwera G, Maes I, Dujardin JC, Requena JM. Evolution and species discrimination according to the Leishmania heat-shock protein 20 gene. Infect Genet Evol. 2013;18:229–37.
Fraga J, Montalvo AM, De Doncker S, Dujardin JC, Van der Auwera G. Phylogeny of Leishmania species based on the heat-shock protein 70 gene. Infect Genet Evol. 2010;10(2):238–45.
Kerr SF. Molecular trees of trypanosomes incongruent with fossil records of hosts. Mem Inst Oswaldo Cruz. 2006;101(1):25–30.
Kerr SF, Merkelz R, Mackinnon C. Further support for a Palaearctic origin of Leishmania. Mem Inst Oswaldo Cruz. 2000;95(4):579–81.
Noyes H. Implications of a Neotropical origin of the genus Leishmania. Mem Inst Oswaldo Cruz. 1998;93(5):657–61.
Noyes HA, Morrison DA, Chance ML, Ellis JT. Evidence for a Neotropical origin of Leishmania. Mem Inst Oswaldo Cruz. 2000;95(4):575–8.
Yurchenko VY, Lukeš J, Jirků M, Zeledon R, Maslov DA. Leptomonas costaricensis sp. n. (Kinetoplastea: Trypanosomatidae), a member of the novel phylogenetic group of insect trypanosomatids closely related to the genus Leishmania. Parasitology. 2006;133:537–46.
Harkins KM, Schwartz RS, Cartwright RA, Stone AC. Phylogenomic reconstruction supports supercontinent origins for Leishmania. Infect Genet Evol. 2016;38:101–9.
Votýpka J, Kostygov AY, Kraeva N, Grybchuk-Ieremenko A, Tesařová M, Grybchuk D, et al. Kentomonas gen. n., a new genus of endosymbiont-containing trypanosomatids of Strigomonadinae subfam. n. Protist. 2014;165(6):825–38.
Schaub GA. The effect of Blastocrithidia triatomae (trypanosomatidae) on the reduviid bug Triatoma infestans: influence of group-size. J Invertebr Pathol. 1990;56(2):249–57.
Votýpka J, Suková E, Kraeva N, Ishemgulova A, Duží I, Lukeš J, et al. Diversity of trypanosomatids (Kinetoplastea: Trypanosomatidae) parasitizing fleas (Insecta: Siphonaptera) and description of a new genus Blechomonas gen. n. Protist. 2013;164(6):763–81.
Runckel C, DeRisi J, Flenniken ML. A draft genome of the honey bee trypanosomatid parasite Crithidia mellificae. PLoS One. 2014;9(4):10.
Schwarz RS, Bauchan GR, Murphy CA, Ravoet J, de Graaf DC, Evans JD. Characterization of two species of trypanosomatidae from the honey bee Apis mellifera: Crithidia mellificae Langridge and McGhee, and Lotmaria passim n. gen., n. sp. J Eukaryot Microbiol. 2015;62(5):567–83.
Svobodová M, Zidková L, Cepicka I, Oborník M, Lukeš J, Votýpka J. Sergeia podlipaevi gen. nov., sp. nov. (Trypanosomatidae, Kinetoplastida), a parasite of biting midges (Ceratopogonidae, Diptera). Int J Syst Evol Microbiol. 2007;57:423–32.
Kostygov AY, Grybchuk-Ieremenko A, Malysheva MN, Frolov AO, Yurchenko V. Molecular revision of the genus Wallaceina. Protist. 2014;165(5):594–604.
de Oliveira SSC, Goncalves DD, Garcia-Gomes AD, Goncalves IC, Seabra SH, Menna-Barreto RF, et al. Susceptibility of Phytomonas serpens to calpain inhibitors in vitro: interference on the proliferation, ultrastructure, cysteine peptidase expression and interaction with the invertebrate host. Mem Inst Oswaldo Cruz. 2017;112(1):31–43.
Hayes PM, Lawton SP, Smit NJ, Gibson WC, Davies AJ. Morphological and molecular characterization of a marine fish trypanosome from South Africa, including its development in a leech vector. Parasit Vectors. 2014;7(1):50.
Sehgal RNM, Jones HI, Smith TB. Host specificity and incidence of Trypanosoma in some African rainforest birds: a molecular approach. Mol Ecol. 2001;10(9):2319–27.
Githure JI, Hendricks L, Schnur L, Kiilu G, Perkins P. Characterization of Crithidia-like organisms isolated from man, animals and sand flies in leishmaniasis-endemic foci in Kenya. East Afr Med J. 1986;50:243–7.
Conchon I, Campaner M, Sbravate C, Camargo EP. Trypanosomatids, other than Phytomonas spp., isolated and cultured from fruit. J Protozool. 1989;36(4):412–4.
Sabbatani S, Calzado AI, Ferro A, Goudiaby AML, Borghi V, Zanchetta GP, et al. Atypical leishmaniasis in an HIV-2-seropositive patient from guinea-bissau. AIDS. 1991;5(7):899–901.
Mebrahtu YB, Lawyer PG, Pamba H, Koech D, Perkins PV, Roberts CR, et al. Biochemical characterization and zymodeme classification of Leishmania isolates from patients, vectors, and reservoir hosts in Kenya. Am J Trop Med Hyg. 1992;47(6):852–92.
Jimenez MI, LopezVelez R, Molina R, Canavate C, Alvar J. HIV co-infection with a currently non-pathogenic flagellate. Lancet. 1996;347(8996):264–5.
Dedet JP, Roche B, Pratlong F, CalesQuist D, Jouannelle J, Benichou JC, et al. Diffuse cutaneous infection caused by a presumed monoxenous trypanosomatid in a patient infected with HIV. Trans R Soc Trop Med Hyg. 1995;89(6):644–6.
Boisseau-Garsaud AM, Cales-Quist D, Desbois N, Jouannelle J, Jouannelle A, Pratlong F, et al. A new case of cutaneous infection by a presumed monoxenous trypanosomatid in the island of Martinique (French West Indies). Trans R Soc Trop Med Hyg. 2000;94(1):51–2.
Pothirat T, Tantiworawit A, Chaiwarith R, Jariyapan N, Wannasan A, Siriyasatien P, et al. First isolation of Leishmania from Northern Thailand: case report, identification as Leishmania martiniquensis and phylogenetic position within the Leishmania enriettii complex. PLoS Negl Trop Dis. 2014;8(12):8.
Hailu A, Musa AM, Royce C, Wasunna M. Visceral leishmaniasis: New health tools are needed. PLoS Med. 2005;2(7):590–4.
Under My Skin: After infection, sleeping sickness parasite subpopulation grows in host skin [http://researchnews.plos.org/2016/08/08/under-my-skin/]
Curtis-Robles R, Wozniak EJ, Auckland LD, Hamer GL, Hamer SA. Combining public health education and disease ecology research: using citizen science to assess Chagas disease entomological risk in Texas. PLoS Negl Trop Dis. 2015;9(12):12.
Gois L, Badaró R, Schooley R, Grassi MFR. Immune response to Leishmania antigens in an AIDS patient with mucocutaneous leishmaniasis as a manifestation of immune reconstitution inflammatory syndrome (IRIS): a case report. BMC Infect Dis. 2015;15(1):38.
Mohammadpour I, Motazedian MH, Handjani F, Hatam GR. Lip leishmaniasis: a case series with molecular identification and literature review. BMC Infect Dis. 2017;17(1):96.
Al-Dwibe H, Gashout A, Morogum AM, El-Zubi S, Amro A. Contact dermatitis-like cutaneous leishmaniasis in a Libyan HIV patient. Parasit Vectors. 2014;7:3.
Pennisi MG, Cardoso L, Baneth G, Bourdeau P, Koutinas A, Miro G et al. LeishVet update and recommendations on feline leishmaniosis. 2015;8:302.
Baneth G, Zivotofsky D, Nachum-Biala Y, Yasur-Landau D, Botero AM. Mucocutaneous Leishmania tropica infection in a dog from a human cutaneous leishmaniasis focus. Parasit Vectors. 2014;7:5.
Ordeix L, Dalmau A, Osso M, Llull J, Montserrat-Sangra S, Solano-Gallego L. Histological and parasitological distinctive findings in clinically-lesioned and normal-looking skin of dogs with different clinical stages of leishmaniosis. Parasit Vectors. 2017;10:8.
Acknowledgements
Not applicable.
Funding
This research was funded by the University of Technology Sydney Chancellor’s fellowship to Dr Joel Barratt.
Availability of data and materials
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
Authors’ contributions
AK conceived, wrote and edited all drafts. JE and DS provided additional edits and suggestions on content. JB provided major suggestions on content, major edits, and approved the final draft. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kaufer, A., Ellis, J., Stark, D. et al. The evolution of trypanosomatid taxonomy. Parasites Vectors 10, 287 (2017). https://doi.org/10.1186/s13071-017-2204-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-017-2204-7