Abstract
Background
The Ehrlichia are obligate intracellular Gram-negative tick-borne bacteria that are important human and animal pathogens. There is a need for assays to rapidly and reliably detect and differentiate the five generally recognized species into groups in a single reaction: E. canis, E. chaffeensis, E. ewingii, E. muris and E. ruminantium.
Methods
We developed primers and probes against the 16S rRNA gene to enable us to reliably detect the five major Ehrlichia spp. in a single FRET-qPCR. We tested the Ehrlichia FRET-qPCR on reference strains and on DNA from the blood of domestic ruminants from five Caribbean islands. The Ehrlichia present were determined using melting point analysis and by sequencing the Ehrlichia FRET-qPCR products as well as those of a nested PCR against the citrate synthase gene (gltA).
Results
Our Ehrlichia FRET-qPCR was negative for the closely related Anaplasma marginale and A. phagocytophilum but gave positive reactions with reference strains of the most generally recognized species and with other less characterized Ehrlichia of domestic ruminants, mainly E. ovina, the Panola Mountain Ehrlichia, and Ehrlichia sp. BOV2010. Melting point analysis revealed 4 distinct groups: E. ruminantium (T m ~55.8 °C); E. chaffeensis and E. ewingii (T m ~57.7 °C); E. canis, E. muris, E. ovina and Ehrlichia sp. BOV 2010 (T m ~62.0 °C); and the Panola Mountain Ehrlichia (T m ~65.5 °C). The detection limit of the FRET-qPCR was ~ 5 gene copies in a reaction and the sequences of the FRET-qPCR products were as expected. With DNA from domestic ruminants from the Caribbean we found 12.2 % (134/1,101) positive: cattle (76/385; 19.7 %), sheep (45/340; 13.2 %) and goats (13/376; 3.5 %). Melting point analysis and sequencing of the FRET-qPCR and nested PCR gltA products showed the Ehrlichia we detected were E. canis or very closely related organisms.
Conclusions
In a single reaction, our Ehrlichia FRET-qPCR can detect the Ehrlichia spp. we studied and differentiate them into four groups. Domestic ruminants in the Caribbean are not uncommonly exposed to Ehrlichia, possibly E. canis or very closely related organisms.
Similar content being viewed by others
Background
Ehrlichia are obligate intracellular Gram-negative tick-borne bacteria that are important animal and human pathogens. There are five generally recognized Ehrlichia spp., mainly E. canis, E. chaffeensis, E. ewingii, E. muris and E. ruminantium [1, 2]. Ehrlichia ruminantium is the most important in domestic ruminants, where it causes heartwater, an acute disease associated with very high mortality (up to 90 %) and extensive economic losses [3]. Although various serological tests for E. ruminantium have been described, in particular ELISAs detecting antibodies to the organism’s major antigenic protein (MAP), inappropriate positive results are not uncommon, probably due to cross-reactivity with other tick-borne Ehrlichia spp. [4–10]. A number of such Ehrlichia that might be responsible for the serological cross-reactivity have been described in domestic ruminants, including E. ovina in a sheep from Turkey [4], E. chaffeensis in goats and cattle in the USA [11, 12], the Panola Mountain Ehrlichia in goats in the USA [9], and Ehrlichia sp. BOV2010/Ehrlichia sp. UFMT-BV in cattle in the Americas [13, 14]. There are also other Ehrlichia that have been reported in domestic ruminants, but stocks are not readily available and their taxonomic status is yet to be confirmed [15]. These include E. ondiri [15], Ehrlichia sp. Omatjenne [16], Ehrlichia sp. Germishuys [16] and an Ehrlichia sp. from Zimbabwe [7].
In a recent study in the Caribbean, inappropriate positive MAP-1B ELISA results for E. ruminantium were reported for domestic ruminants from four of the seven islands studied [10]. These inappropriate positive reactions were thought to be due to infections with other Ehrlichia spp., and the presence of these organisms made serological testing for E. ruminantium unreliable in the Caribbean, as has been shown to be the case in Africa [17]. Being able to reliably detect E. ruminantium is important as it is not only a serious threat to local livestock production, but also to animals on the American mainland [18]. To further investigate ehrlichioses in domestic ruminants in the Caribbean, we developed a generic Ehrlichia FRET-qPCR that would enable us in a single reaction to specifically and reliably detect the major Ehrlichia spp. and differentiate them into groups. The development and validation of this PCR and its use to screen domestic ruminants in the Caribbean for Ehrlichia spp. is described below.
Methods
Blood samples
Whole blood samples (n = 1,101) in EDTA were collected from apparently healthy domestic ruminants (cattle, sheep and goats) on Montserrat (n = 77), St. Kitts (n = 373), Grenada (n = 140), Nevis (n = 262) and Dominica (n = 249) as described previously [10, 19] (Table 1). Aliquots of 200 μl were frozen at −20 °C until DNA was extracted for PCR studies. Ethical Approval: All work in this study was reviewed and approved by the Institutional Animal Care and Use Committee of Ross University School of Veterinary Medicine. Owners of the animals provided consent for blood samples to be collected.
Ehrlichia strains
As positive controls we used the five major Ehrlichia spp., mainly E. ruminantium, E. canis, E. chaffeensis, E. ewingii and E. muris. We also tested Ehrlichia that were available to us and have been previously reported to occur in ruminants, mainly E. ovina [4], Ehrlichia sp. BOV2010 [13] and the Panola Mountain Ehrlichia [9]. We used DNA extracted in previous studies from E. ruminantium [10] and E. canis [20], DNA extracted as described below from tissue cultures of E. canis (Oklahoma) and E. chaffeensis (Arkansas) (supplied by Gregory Dasch, Centers for Disease Control, Atlanta), from blood stabilates (E. ovina and Ehrlichia sp. BOV2010), and from an Amblyomma variegatum positive for the Panola Mountain Ehrlichia by PCR (unpublished data). We also used plasmids that were created to contain an appropriate portion of the 16S rRNA gene of E. ewingii and E. muris using the pIDTSMART cloning vector (Integrated DNA Technologies, Coralville, IA, USA) and linearization with HindIII (Promega, Madison, WI, USA).
To test the specificity of our PCR, we tested DNAs extracted from blood of cattle verified to be infected with A. marginale (identical nucleotide 16S rRNA sequences with CP006847) and A. phagocytophilum (identical 16S rRNA sequences with KJ782389).
DNA extraction
The High-Pure PCR Template Preparation Kit (Roche Molecular Biochemicals, Indianapolis, IN, USA) was used according to the manufacturer’s instructions to extract total nucleic acids from the samples (200 μl). The extracted DNAs were eluted in 200 μl elution buffer and stored at −80 °C.
Development of a generic Ehrlichia FRET-qPCR
Primers and probes
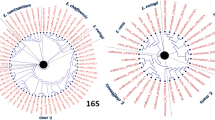
The 16S rRNA sequences for the five major Ehrlichia spp. and those reliably reported in domestic ruminants, five Anaplasma spp., and six related bacteria were obtained from GenBank: E. canis (EU178797, GU810149), E. ruminantium (CR925678, DQ647616, U03776, U03777), E. chaffeensis (AF147752, U60476), E. ewingii (M73227, U96436), E. muris (AB013008, AB196302), E. ovina (AF318946), Ehrlichia sp. BOV 2010 (HM486680), the Panola Mountain Ehrlichia (DQ324367); A. equi (AF172167), A. platys (M82801), A. phagocytophilum (AY055469), A. bovis (HQ913646), A. marginale (AF309866, AF414873); Bartonella henselae (AY513504); Rickettsia rickettsii (L36217), Neorickettsia helminthoeca (U12457), Neorickettsia risticii (NR029162); Coxiella burnetii (D89798), and Eperythrozoon sp. (FR869692) (Fig. 1). The sequences were aligned and regions were identified for primers and probes based on the conserved and variable areas of the alignments. The forward primer (5′-GAGGATTTTATCTTTGTATTGTAGCTAAC-3′), reverse primer (5′-TGTAAGGTCCAGCCGAACTGACT-3′) and fluorescein probe (5′-ACGCGAAAAACCTTACCACTTTTTGAC-6-FAM-3′) we selected had identical sequences in all the Ehrlichia. The LCRed 640 probe (5′-LCRred640-GAAGGTCGTATCCCTCTTAACAGG-phos-3′) was identical to the Panola Mountain Ehrlichia but had one nucleotide mismatch with E. canis, E. muris, E. ovina and Ehrlichia sp. BOV 2010, two mismatches with E. ewingii and E. chaffeensis, and three mismatches with E. ruminantium (Fig. 1). In contrast, the primers and probes had multiple mismatches (19–57) with Anaplasma spp. and other related bacteria (Fig. 1). When we used the BLAST to compare the primers and probes we developed against all sequences available on GenBank, we found they reliably detected the Ehrlichia spp. against which they were designed and that the nucleotide polymorphisms we used in the probes for the different species were highly conserved.
Alignment of the primers and probes of the generic Ehrlichia FRET-qPCR with the 16S rRNA gene sequences of Ehrlichia spp. and related genera/species. The sequences of the upstream/downstream primers and the fluorescein/LCRed 640 probes are shown at the top of the boxes. The upstream primer and two probes were used as the indicated sequences while the downstream primer was used as an antisense oligonucleotide. The 6-FAM label was attached directly to the 3-terminal nucleotide of the fluorescein probe and the LCRed 640 fluorescein label was added via a linker to the 5′-end of the LCRed 640 probe. Dots indicate nucleotides identical to the primers and probes, and dashes denote the deletion of a nucleotide. Both of the primers and the fluorescein probe had 100 % identity with all Ehrlichia spp. while the LCRed probe had 0, 1, 2 or 3 nucleotide mismatches. The primers and probes had multiple mismatches with other related organisms
Thermal cycling and melting curve analysis
High-resolution melting curve analysis following PCR was performed on a Roche Light-Cycler 480-II platform as described before [21, 22]. Each reaction was performed in a 20 μL final volume containing 10 μL of extracted DNA. Thermal cycling consisted of 1 activation cycle of 5 min at 95 °C followed by 45 fluorescence acquisition cycles consisting of 10 s at 95 °C, 15 s at 58 °C, and 15 s at 72 °C. Melting curve analysis was performed by monitoring fluorescence between 45 °C and 80 °C after 30 s at 95 °C. Data were analyzed as 640 nm: 530 nm (F4/F1) fluorescence ratios, and the first derivative of F4/F1 (−d(F4/F1)/dt) was evaluated (Fig. 2). The T m value is influenced not only by nucleotide mismatches but also the types of nucleotides and GC percentage of the probes.
Composite of melting curves obtained with the generic Ehrlichia FRET-qPCR performed on various Ehrlichia species. The nucleotide mismatches between amplicons of the various species and the LCRed-640 probe we designed (Fig. 1) enabled us to distinguish four groups of Ehrlichia based on their previously determined T m: Panola Mountain Ehrlichia ~65.5 °C (green line); E. canis, E. muris, E. ovina and Ehrlichia sp. BOV2010/Ehrlichia sp. UMFG-EV ~62.0 °C (blue line); E. chaffeensis and E. ewingii (red line) ~57.6 °C; E. ruminantium ~55.8 °C (black line). No amplification peak was seen with A. marginale DNA (grey line)
Sensitivity
For quantitative standards we used amplified DNA of E. canis identified in a previous study [20]. These E. canis DNA amplification products were confirmed by nucleotide sequencing (GenScript, Nanjing, Jiangsu, China) before being gel purified with a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA) and quantified using the PicoGreen DNA fluorescence assay (Molecular Probes, Eugene, OR). The molarity of the E. canis DNA was estimated using the calculated molecular mass of the amplicons [23] and dilutions made to give solutions containing 10,000, 1,000, 100, 10, and 1 gene copies/μl in T10E0.1 buffer which were used as quantitative standards.
Specificity
The specificity of the positive control PCRs with the five widely recognized Ehrlichia spp. (DNAs of E. canis, E. chaffeensis and E. ruminantium, and plasmids representing E. ewingii and E. muris) were confirmed by electrophoresis of amplicons through 1.5 % MetaPhor agarose gels, purification using the QIAquick PCR Purification Kit (Qiagen, Valencia, CA, USA) and sequencing of both DNA strands using the appropriate forward and reverse primers (GenScript, Jiangsu, Nanjing, China). No reaction products were obtained when our generic Ehrlichia FRET-qPCR was performed with DNAs of A. marginale or A. phagocytophilum.
Nested PCR for the citrate synthase gene of Ehrlichia
To amplify the citrate synthase gene (gltA) of Ehrlichia spp., we carried out nested PCRs (outside primers: EHRCS-131F and EHRCS-1226R, and inside primers: EHRCS-754F and EHRCS-879R, which amplify 1,108 and 126 bp sections of the gene, respectively) as described previously [9]. The PCR products we obtained were verified by gel electrophoresis, purified using the QIAquick PCR Purification Kit (Qiagen, Valencia, CA, USA) and sequenced (GenScript, Jiangsu, Nanjing, China).
Results
Development of a generic Ehrlichia FRET-qPCR
The generic Ehrlichia FRET-qPCR we established produced amplicons with each of the five well recognized Ehrlichia spp. that we tested, mainly E. ruminantium, E. chaffeensis, E. ewingii, E. canis and E. muris. The FRET-qPCR was also positive with DNA from E. ovina, the Panola Mountain Ehrlichia, and Ehrlichia sp. BOV 2010. Sequences of the amplification products were as expected for each organism (results not shown). No products were obtained when the generic Ehrlichia FRET-qPCR was performed with DNA from A. marginale and A. phagocytophilum (results not shown). Further, we obtained no amplification products when we tested DNA from 60 of the cattle from St Kitts that were seropositive for Anaplasma marginale which is endemic and highly prevalent in the Caribbean (results not shown) (Kelly PJ, unpublished data).
Melting curve analysis enabled us to identify 4 distinct groups of Ehrlichia based on their T m: E. ruminantium ~55.8 °C; E. chaffeensis and E. ewingii ~57.7 °C; E. canis, E. muris, E. ovina and Ehrlichia sp. BOV 2010 ~ 62.0 °C; the Panola Mountain Ehrlichia ~65.5 °C (Fig. 2). No reaction products or melting peaks were found with the positive DNAs of A. marginale and A. phagocytophilum. When we tested around 300 copies of E. ruminantium, E. chaffeensis, Ehrlichia sp. BOV 2010 and the Panola Mountain Ehrlichia/μL in a single reaction, the Ehrlichia FRET-qPCR revealed 4 distinct melting curves with temperatures identical to those found with individual FRET-qPCRs of the agents.
With the quantitative standards developed using purified E. canis DNA, we determined that the detection limit of the generic Ehrlichia FRET-qPCR was ~5 copies of the 16S rRNA gene per PCR.
Prevalence of Ehrlichia spp. in domestic ruminants from five Caribbean islands
Of the 1,101 blood samples we examined, 134 (12.2 %) were positive for Ehrlichia spp. in our generic Ehrlichia FRET-qPCR (Table 1). Cattle were most commonly positive (19.7 %; 76/385), followed by sheep (13.2 %; 45/340) and goats (3.5 %; 13/376). The average 16S rRNA copy number in the Ehrlichia-positive samples was 231 per μl of blood. All positive reactions had a T m of ~62.0 °C and sequencing of seven animals’ 16S rRNA amplicons showed the organisms we detected were 98–100 % identical with strains of E. canis from Turkey (Kutahya:AY621071) and the Philippines (D28A: JN121380), Ehrlichia sp. BOV2010 (HM486680) from Canada and the Ehrlichia UFMG-EV (JX629805) from Brazil [24]. They also had 98–100 % similarity with E. ovina and the Ehrlichia sp. Germishuys (U54805) which has a 16S rRNA sequence 99.9 % identical to E. canis [16]. Sequencing of 15 of the nested gltA products we obtained revealed eight (Group 1; Table 2) had 98 % identity with the Ehrlichia sp. BOV2010 (JN673762) and the Ehrlichia sp. UFMG-EV (JX629807), 5 (Groups 3 and 4; Table 2) had 96 % similarity with E. canis from the US (Jake; NC007354) and Italy (AY647155), one (Group 5; Table 2) had 99 % identity with an Ehrlichia identified in a cattle tick in Africa (AF311965) [25], and one (Group 2; Table 2) had 95 % similarity with the Ehrlichia sp. BOV2010 (JN673762) and the Ehrlichia sp. UFMG-EV (JX629807). All the groups generally shared least similarity with E. ruminantium and the Panola Mountain Ehrlichia.
The 1,015 bp gltA sequence we obtained for E. ovina and deposited in GenBank (KP719095) was 99.9 % (2 mismatches) identical to that of E. canis from Italy (AY647155).
Discussion
The generic Ehrlichia FRET-qPCR we developed proved to be both specific and sensitive in detecting Ehrlichia spp. in controlled experiments. In a single reaction it reliably detected the five commonly recognized Ehrlichia spp. we used in our experiments as well as less well characterized Ehrlichia which have been found in domestic ruminants and are available for study [26]. The specificity of the PCR was shown by its failure to detect representatives of the closely related Anaplasma genus, A. marginale and A. phagocytophilum, and the fact that all the positive reaction products had sequences that were closest to Ehrlichia spp. When tested against dilutions of E. canis, the sensitivity of the generic Ehrlichia FRET-qPCR was high, detecting as few as 5 copies of the 16S rRNA gene in a reaction [27].
The 16S rRNA gene we detected in our generic Ehrlichia FRET-qPCR is a common target for PCRs for Ehrlichia spp. as its nucleotide sequence is highly conserved in the genus. By systematically aligning the sequences of the main Ehrlichia spp. and closely related organisms, we were able to identify a highly conserved region of the 16S rRNA gene against which we developed specific primers that only amplified Ehrlichia spp. and not organisms from related genera. Further, the region of the 16S rRNA gene we selected for our LCRed 640 probe had nucleotide mismatches between the major Ehrlichia spp. which enabled us to differentiate the organisms into groups by melting point analysis (Fig. 1).
When we tested our generic Ehrlichia FRET-qPCR against known Ehrlichia spp. it detected all the organisms in a single reaction and also differentiated the species to a large extent. Using high-resolution melting point analysis we were able to clearly differentiate E. ruminantium, the Panola Mountain Ehrlichia, a group containing E. chaffeensis and E. ewingii, and a group containing E. muris as well as E. canis and organisms closely related to it. The groupings appear to be largely serendipitous, rather than of taxonomic significance, as molecular studies have shown E. muris is more closely related to E. chaffeensis than to E. canis [26]. Similarly, E. ewingii is closer to E. canis [27] or the Panola Mountain Ehrlichia [9] than to E. chaffeensis.
When we applied our generic Ehrlichia FRET-qPCR to DNA from whole blood collected from domestic ruminants from five Caribbean islands, we identified relatively high prevalences of infections (12 %) with Ehrlichia spp. that were not E. ruminantium. Of note is the fact these generic Ehrlichia FRET-qPCR positive animals had previously tested negative for antibodies to E. ruminantium in a MAP-1B ELISA [20]. This test not only detects antibodies to E. ruminantium [6] but also to the Panola Mountain Ehrlichia [9], and E. canis and E. chaffeensis [6]. It seems unlikely, then, that the animals we found positive in our generic Ehrlichia FRET-qPCR had been infected with these agents. While there are no data for Ehrlichia sp. BOV2010 and Ehrlichia sp. UMFG-EF, sera from animals infected with E. ovina do not give positive MAP-1B ELISA reactions [6] and these, or closely related organisms, seem most likely to have been detected by our generic Ehrlichia FRET-qPCR in the seronegative animals. Also of note is that none of the animals that had previously been found to be positive in MAP-1B ELISAs [10] were positive in our generic Ehrlichia FRET-qPCR. Most of these animals, however, were only very weakly positive in the MAP-1B ELISA suggesting they had residual antibody titers following clearance of infections, or that the infecting Ehrlichia spp. did not generate a substantial humoral response. Further studies are underway in our laboratories to clarify the position.
Melting point analysis and sequencing suggested that the Ehrlichia we identified with our generic Ehrlichia FRET-qPCR, utilizing the 16S rRNA gene, were E. canis or closely related organisms. The 16S rRNA gene is highly conserved in E canis, being 99.4–100 % identical between strains [28, 29], and hence a reliable way of identifying isolates. Although we sequenced only a relatively short segment of the gene (210 bp), the Ehrlichia spp. we identified with our generic Ehrlichia FRET-qPCR had 100 % homology with E. canis sequences in GenBank.
We found only relatively small numbers of organisms in the blood samples we studied (average copy number 231, median 9.5) with our generic Ehrlichia FRET-qPCR, most likely because we were detecting chronic subclinical infections but also perhaps because we were detecting infections in accidental and unsuitable hosts. The low copy numbers in our samples were also evident from the results of our gltA gene PCRs where we only found positive results after nesting. The gltA gene has also been shown to be highly conserved in E. canis (over 99 %) [30], but it has greater interspecies variability than the 16S rRNA gene which might make it more useful for differentiating Ehrlichia species [9, 31]. The sequences we obtained for our nested gltA, however, were consistent with the 16S rRNA gene findings that the organisms present in the Caribbean domestic ruminants we studied were E. canis or closely related species.
We would note that, because of low copy numbers in our samples, the sequences we obtained from our nested gltA PCR were with the internal primers and thus relatively short (126 bp). These internal primers, however, amplify a hypervariable region of the gltA which enables accurate discrimination of species and strains. When we compared the sequences we obtained with others in GenBank we found that, consistent with comparisons of our 16S rRNA gene sequences, the organisms present in the Caribbean domestic ruminants were closest to E. canis or closely related organisms.
Although E. canis is best known as a very common dog pathogen around the world, including in the Caribbean [20, 32], infections have also been described in humans [33] and cats [34]. There is a growing belief that E. canis has a wider host range than previously thought [1, 14], and our findings are largely consistent with this idea. Of further note is that a number of Ehrlichia that appear to be closely related to E. canis, possibly even strains of this organism, have been reported in domestic ruminants. E. ovina (AF318946) was first recovered from a sheep in Turkey and subsequently caused illness in splenectomized Dutch sheep [4]. More recently it has been found to have an identical 16S rRNA sequence to E. canis in dogs from Turkey (Kutahya strain; AY621071) [35] and Venezuela (VHE strain; AF373612) [36]. In our study, E. ovina had a T m and 16S rRNA sequence identical to that of the Oklahoma strain of E. canis (NR_118741) and of local Caribbean strains we found. Further, E. ovina reacted with primers for the gltA of Ehrlichia spp. and produced a 1015 bp sequence that contained only 2 mismatches with E. canis from Italy (AY647155). These findings provide further support for the proposal that E. ovina is a strain of E. canis [1, 34].
Recent studies have identified 3 novel cattle-related strains of Ehrlichia: in Canada, the Ehrlichia sp. BOV 2010 [13]; and in Brazil, the Ehrlichia sp. UFMG-EV in Rhipicephalus microplus hemolymph [37] and the Ehrlichia sp. UFMT-BV in cattle [1]. Molecular studies have shown these organisms are very similar to one another and that they probably evolved from a highly divergent and variable clade within E. canis [38]. The phenotypic and genotypic differences the strains have with E. canis have been ascribed to the organisms adapting to their new hosts, ruminants, and their new tick vectors. More detailed genomic and transmission studies might provide justification for the organism being classified as a distinct species, E. mineirensis [38].
Conclusions
In conclusion, the Ehrlichia FRET-qPCR we developed proved sensitive and specific in detecting the most recognized Ehrlichia spp. of ruminants in a single reaction. Further, using melting point analysis we could differentiate the organisms into four groups comprising E. ruminantium; E. chaffeensis and E. ewingii; E. canis and closely related organisms such as E. ovina and Ehrlichia sp. BOV2010/Ehrlichia sp. UFMG-EV; and the Panola Mountain Ehrlichia. When we used the generic Ehrlichia FRET-qPCR on DNA from the blood of Caribbean domestic ruminants, we found a relatively high percentage (12.2 %) were positive. Melting point analysis showed the Ehrlichia in the Caribbean domestic ruminants were most similar to organisms in the group comprising E. canis and closely related species.
References
Yu XJ, McBride JW, Walker DH. Restriction and expansion of Ehrlichia strain diversity. Vet Parasitol. 2007;143:337–46.
Esemu SN, Ndip LM, Ndip RN. Ehrlichia species, probable emerging human pathogens in sub-Saharan Africa: environmental exacerbation. Rev Environ Health. 2011;26:269–79.
Mahan SM, Smith GE, Kumbula D, Burridge MJ, Barbet AF. Reduction in mortality from heartwater in cattle, sheep and goats exposed to field challenge using an inactivated vaccine. Vet Parasitol. 2001;97:295–308.
Jongejan F, de Vries N, Nieuwenhuijs J, Van Vliet AH, Wassink LA. The immunodominant 32-kilodalton protein of Cowdria ruminantium is conserved within the genus Ehrlichia. Rev Elev Med Vet Pays Trop. 1993;46:145–52.
Kelly PJ, Matthewman LA, Mahan SM, Semu S, Peter T, Mason PR, et al. Serological evidence for antigenic relationships between Ehrlichia canis and Cowdria ruminantium. Res Vet Sci. 1994;56:170–4.
van Vliet AH, van der Zeijst BA, Camus E, Mahan SM, Martinez D, Jongejan F. Use of a specific immunogenic region on the Cowdria ruminantium MAP1 protein in a serological assay. J Clin Microbiol. 1995;33:2405–10.
Savadye DT, Kelly PJ, Mahan SM. Evidence to show that an agent that cross reacts serologically with Cowdria ruminantium in Zimbabwe is transmitted by ticks. Exp Appl Acarol. 1998;22:111–22.
Kakono O, Hove T, Geysen D, Mahan S. Detection of antibodies to the Ehrlichia ruminantium MAP1-B antigen in goat sera from three communal land areas of Zimbabwe by an indirect enzyme-linked immunosorbent assay. Onderstepoort J Vet Res. 2003;70:243–9.
Loftis AD, Reeves WK, Spurlock JP, Mahan SM, Troughton DR, Dasch GA, et al. Infection of a goat with a tick-transmitted Ehrlichia from Georgia, USA, that is closely related to Ehrlichia ruminantium. J Vector Ecol. 2006;31:213–23.
Kelly PJ, Lucas H, Yowell C, Beati L, Dame J, Urdaz-Rodriguez J, et al. Ehrlichia ruminantium in Amblyomma variegatum and domestic ruminants in the Caribbean. J Med Entomol. 2011;48:485–8.
Dugan VG, Little SE, Stallknecht DE, Beall AD. Natural infection of domestic goats with Ehrlichia chaffeensis. J Clin Microbiol. 2000;38:448–9.
delos Santos JR, Oglesbee M, Rikihisa Y, Stich RW. Pathologic evidence of ehrlichiosis in calves inoculated with Ehrlichia chaffeensis. Ann N Y Acad Sci. 2008;1149:103–6.
Gajadhar AA, Lobanov V, Scandrett WB, Campbell J, Al-Adhami B. A novel Ehrlichia genotype detected in naturally infected cattle in North America. Vet Parasitol. 2010;173:324–9.
Aguiar DM, Ziliani TF, Zhang X, Melo AL, Braga IA, Witter R, et al. A novel Ehrlichia genotype strain distinguished by the TRP36 gene naturally infects cattle in Brazil and causes clinical manifestations associated with ehrlichiosis. Ticks Tick Borne Dis. 2014;5:537–44.
Sumption KJ, Scott GR. Lesser-known rickettsias infecting livestock. In: Coetzer JAW, Tustin RC, editors. Infectious Diseases of Livestock. Cape Town: ABC Press; 2005. p. 536–49.
Allsopp BA, Allsopp MT, Du Plessis JH, Visser ES. Uncharacterized Ehrlichia spp. may contribute to clinical heartwater. Ann N Y Acad Sci. 1996;791:17–23.
Vachiéry N, Marcelino I, Martinez D, Lefrançois T. Opportunities in diagnostic and vaccine approaches to mitigate potential heartwater spreading and impact on the American mainland. Dev Biol (Basel). 2013;135:191–200.
Burridge MJ, Simmons LA, Peter TF, Mahan SM. Increasing risks of introduction of heartwater onto the American mainland associated with animal movements. Ann N Y Acad Sci. 2002;969:269–74.
Kelly P, Lucas H, Randolph C, Ackerson K, Blackburn J, Dark M. Efficacy of slow-release tags impregnated with aggregation-attachment pheromone and deltamethrin for control of Amblyomma variegatum on St. Kitts, West Indies. Parasit Vectors. 2014;7:182.
Kelly PJ, Xu C, Lucas H, Loftis A, Abete J, Zeoli F, et al. Ehrlichiosis, babesiosis, anaplasmosis and hepatozoonosis in dogs from St. Kitts, West Indies. PLoS One. 2013;8, e53450. doi:10.1371/journal.pone.0053450.
Wang C, Johnson CM, Ahluwalia SK, Chowdhury E, Li Y, Gao D, et al. Dual-emission fluorescence resonance energy transfer (FRET) real-time PCR differentiates feline immunodeficiency virus subtypes and discriminates infected from vaccinated cats. J Clin Microbiol. 2010;48:1667–72.
Zhang J, Wei L, Kelly P, Freeman M, Jaegerson K, Gong J, et al. Detection of Salmonella spp. using a generic and differential FRET-PCR. PLoS One. 2013;8, e76053. doi:10.1371/journal.pone.0076053.
Yang Y, Mao Y, Kelly P, Yang Z, Luan L, Zhang J, et al. A pan-Theileria FRET-qPCR survey for Theileria spp. in ruminants from nine provinces of China. Parasit Vectors. 2014;7:413.
Cruz AC, Zweygarth E, Ribeiro MF, da Silveira JA, de la Fuente J, Grubhoffer L, et al. New species of Ehrlichia isolated from Rhipicephalus (Boophilus) microplus shows an ortholog of the E. canis major immunogenic glycoprotein gp36 with a new sequence of tandem repeats. Parasit Vectors. 2012;5:291.
Parola P, Inokuma H, Camicas JL, Brouqui P, Raoult D. Detection and identification of spotted fever group Rickettsiae and Ehrlichiae in African ticks. Emerg Infect Dis. 2001;7:1014–7.
Wen B, Rikihisa Y, Mott J, Fuerst PA, Kawahara M, Suto C. Ehrlichia muris sp. nov., identified on the basis of 16S rRNA base sequences and serological, morphological, and biological characteristics. Int J Syst Bacteriol. 1995;45:250–4.
Sumner JW, Storch GA, Buller RS, Liddell AM, Stockham SL, Rikihisa Y, et al. PCR amplification and phylogenetic analysis of groESL operon sequences from Ehrlichia ewingii and Ehrlichia muris. J Clin Microbiol. 2000;38:2746–9.
Huang CC, Hsieh YC, Tsang CL, Chung YT. Sequence and phylogenetic analysis of the gp200 protein of Ehrlichia canis from dogs in Taiwan. J Vet Sci. 2010;11:333–40.
Hsieh YC, Lee CC, Tsang CL, Chung YT. Detection and characterization of four novel genotypes of Ehrlichia canis from dogs. Vet Microbiol. 2010;146:70–5.
Marsilio F, Di Martino B, Meridiani I, Bianciardi P. Direct identification of Ehrlichia canis by a novel polymerase chain reaction method and molecular analysis of the citrate synthase (gltA) gene from various Italian strains. J Vet Diagn Invest. 2006;18:215–7.
Inokuma H, Brouqui P, Drancourt M, Raoult D. Citrate synthase gene sequence: a new tool for phylogenetic analysis and identification of Ehrlichia. J Clin Microbiol. 2001;39:3031–9.
Loftis AD, Kelly PJ, Freeman MD, Fitzharris S, Beeler-Marfisi J, Wang C. Tick-borne pathogens and disease in dogs on St. Kitts, West Indies. Vet Parasitol. 2013;196:44–9.
Perez M, Bodor M, Zhang C, Xiong Q, Rikihisa Y. Human infection with Ehrlichia canis accompanied by clinical signs in Venezuela. Ann N Y Acad Sci. 2006;1078:110–7.
Braga IA, dos Santos LG, Melo AL, Jaune FW, Ziliani TF, Girardi AF, et al. Hematological values associated to the serological and molecular diagnostic in cats suspected of Ehrlichia canis infection. Rev Bras Parasitol Vet. 2013;22:470–4.
Unver A, Rikihisa Y, Borku K, Ozkanlar Y, Hanedan B. Molecular detection and characterization of Ehrlichia canis from dogs in Turkey. Berl Munch Tierarztl Wochenschr. 2005;118:300–4.
Unver A, Perez M, Orellana N, Huang H, Rikihisa Y. Molecular and antigenic comparison of Ehrlichia canis isolates from dogs, ticks, and a human in Venezuela. J Clin Microbiol. 2001;39:2788–93.
Cruz AC, Zweygarth E, Ribeiro MF, da Silveira JA, de la Fuente J, Grubhoffer L, et al. New species of Ehrlichia isolated from Rhipicephalus (Boophilus) microplus shows an ortholog of the E. canis major immunogenic glycoprotein gp36 with a new sequence of tandem repeats. Parasit Vectors. 2012;5:291.
Cabezas-Cruz A, Valdés JJ, de la Fuente J. The glycoprotein TRP36 of Ehrlichia sp. UFMG-EV and related cattle pathogen Ehrlichia sp. UFMT-BV evolved from a highly variable clade of E. canis under adaptive diversifying selection. Parasit Vectors. 2014;7:584.
Acknowledgments
This project was supported by grants from the National Natural Science Foundation of China (NO: 31272575), the Priority Academic Program Development of Jiangsu Higher Education Institutions, Yangzhou, Jiangsu, P. R. China and the Ross University School of Veterinary Medicine. We thank Dr. Gregory Dasch (CDC, Atlanta) for the E. chaffeensis and E. canis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WC, KP and ZJ participated in the design of the study and performed the statistical analysis. ZJ, GW and XC carried out the experiments. WL, JF and LA participated in the samples’ preparation. WC, KP and ZJ drafted the manuscript. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, J., Kelly, P., Guo, W. et al. Development of a generic Ehrlichia FRET-qPCR and investigation of ehrlichioses in domestic ruminants on five Caribbean islands. Parasites Vectors 8, 506 (2015). https://doi.org/10.1186/s13071-015-1118-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-1118-5