Abstract
MicroRNAs are a class of endogenous non-coding RNAs that play a vital role in post-transcriptional gene regulation in eukaryotic cells. In plants and animals, miRNAs are implicated in diverse roles ranging from immunity against viral infections, developmental pathways, molecular pathology of cancer and regulation of protein expression. However, the role of miRNAs in the unicellular model green alga Chlamydomonas reinhardtii remains unclear. The mode of action of miRNA-induced gene silencing in C. reinhardtii is very similar to that of higher eukaryotes, in terms of the activation of the RNA-induced silencing complex and mRNA targeting. Certain studies indicate that destabilization of mRNAs and mRNA turnover could be the major possible functions of miRNAs in eukaryotic algae. Here, we summarize recent findings that have advanced our understanding of miRNA regulatory mechanisms in C. reinhardtii.
Similar content being viewed by others
Background
MicroRNAs (miRNAs) are endogenous non-coding RNAs approximately 21–24 nucleotides (nt) in length with significant roles in post-transcriptional gene regulation. MiRNAs were first identified in 1993 in Caenorhabditis elegans and subsequently in Arabidopsis thaliana, Drosophila, mouse, human and other multicellular organisms [1,2,3,4,5,6,7]. MiRNAs were not described in unicellular Chlamydomonas reinhardtii until 2007, much later than the initial reports in animals and plants [8, 9]. In animals, mature miRNAs are embedded in the duplexed stem loop structures of primary miRNAs (pri-miRNAs) and are excised by the ribonuclease III (RNase III) enzyme Drosha [10, 11] accompanied by the double-stranded RNA-binding domain (dsRBD) protein Pasha and Ars2 [12, 13]. The precursor miRNA (pre-miRNA) is subsequently exported from the nucleus to the cytoplasm by Exportin 5 [14, 15] and processed into a miRNA/miRNA* duplex by another RNase III enzyme, Dicer. One strand of the duplex is the mature miRNA, which is loaded onto the RNA-induced silencing complex (RISC) [16], whose core component is an Argonaute protein (AGO) [17] (Fig. 1a). Although plant miRNA biogenesis also involves two cleavage steps, it differs in some important ways from the process in animals (Fig. 1a). First, the two precise cleavage steps of plant miRNA biogenesis are executed in the nucleus by the same RNase III enzyme, Dicer-like 1 (DCL1), with the assistance of HYL1 and SE [18,19,20,21,22,23,24,25,26]. Another difference is that plant miRNAs are typically 21 nt in length, while animal miRNAs are longer, generally 22–23 nt in length. However, the critical difference is that after the two cleavage steps, plant miRNAs are modified by HEN1 methylation at the 3′ end, which protects them from degradation [27,28,29,30,31,32] (Fig. 1b). In contrast, the modification by HEN1 is uncommon for animal miRNAs. To date, CrDCL3, assisted by DUS16, is the only RNase III enzyme known to cleave pri-miRNAs to generate miRNA duplexes in unicellular C. reinhardtii [33,34,35]. Whether the cleavage of Chlamydomonas miRNAs occurs in the nucleus or in the cytoplasm is still unknown (Fig. 1c).
A schematic of canonical miRNA biogenesis in animals, plants and C. reinhardtii. Mature miRNAs are indicated in red, and miRNA* strands are in black. Homologs with similar functions are shown in the same color. Gray areas in the C. reinhardtii panel indicate unknown factors or processes. a In animals, miRNA genes (MIR) are embedded within the introns of protein-coding genes (Gene A, for example) and transcribed by RNA polymerase II (Pol II). Canonical animal miRNAs are processed by the nuclear RNase III enzyme Drosha, in cooperation with Pasha and Ars2. The precursor miRNA (pre-miRNA) is exported from the nucleus to the cytoplasm by Export5, and it is subsequently cut into a miRNA/miRNA* duplex with 2 nt 3′ overhangs by Dicer, acting together with Loqs. One strand of the duplex is degraded, and the other mature miRNA is loaded onto RISC, whose core component is an AGO protein. b In plants, MIR genes are embedded within the noncoding sequences between protein-coding genes (Gene A and Gene B, for example) and are also transcribed by Pol II. Canonical plant miRNAs are produced by the nuclear RNase III Dicer-like1 (DCL1), assisted by HYL1/DRB1 and SERRATE (SE). DCL1 is responsible for both steps of miRNA processing to produce the miRNA/miRNA* duplex, which is then transported to the cytoplasm by HASTY. The miRNA/miRNA* duplex undergoes 2′-O-methylation modification by HEN1 at the 3′ end. One strand of the methylated duplex is degraded, and the other mature miRNA is loaded onto RISC, whose core component is an AGO protein. Plant miRNAs that are not methylated are subject to 3′ end uridylation by the poly(U) polymerase HESO1, resulting in subsequent degradation. c In Chlamydomonas, MIR genes are embedded within the introns of protein-coding genes (Gene A, for example) and transcribed by Pol II, similar to animal MIR. Chlamydomonas miRNAs are processed by the nuclear RNase III enzyme CrDCL3, in cooperation with DUS16. MUT68 is involved in 3′ end uridylation of miRNAs and may mediate miRNA degradation in cooperation with RRP6. The export protein responsible for transporting Chlamydomonas miRNAs from the nucleus to the cytoplasm is unknown. Whether mature miRNAs in C. reinhardtii are methylated is also presently unknown (gray areas in the diagram)
Mature miRNAs are loaded onto AGOs to activate RISCs, which then cleave the target mRNA or prevent translation through the complementarity of miRNAs to their target mRNAs [17, 36,37,38]. Plant miRNAs require near-perfect complementarity to direct the cleavage of target mRNAs [2, 3, 39]. In contrast, animal miRNAs only require complementarity of the seed sequence and hence a broader range of targets, with one animal miRNA targeting several mRNAs. Animal miRNAs usually inhibit the translation of target proteins [40, 41]. As one example in Chlamydomonas, complementary pairing with the seed sequence of the miRNA cre-miR1174.2 was found to be sufficient to trigger repression of target transcripts [42]. Nevertheless, it remains unclear whether the mode of action of miRNA in Chlamydomonas is more similar to those of animal miRNAs or plant miRNAs. Key factors involved in miRNA biogenesis and regulation identified in animals and higher plants are listed in Table 1. The corresponding factors and processes in unicellular C. reinhardtii are not as well understood, but in recent years, some progress has been made towards characterizing miRNA regulatory mechanisms in this species.
Artificial miRNA (amiRNA) technology was developed to knock down the expression of specific target genes. Generally, an endogenous miRNA precursor is used as the backbone to construct a highly specific and high-throughput amiRNA system [43,44,45,46,47,48]. Short tandem target mimic (STTM) technology is another approach used in plants and animals to suppress the functions of specific miRNAs [49,50,51,52,53]. In C. reinhardtii, however, there have not been any reports in which STTM technology was successfully applied.
In this brief review, we summarize findings from previous studies about miRNA biogenesis and modes of action in plants, animals and unicellular C. reinhardtii, including our own research findings, to shed light on the evolutionary divergence of miRNA regulatory mechanisms between multicellular and unicellular organisms.
Discovery and biogenesis of miRNAs
Large-scale discovery of miRNAs
In the first report of miRNAs, the heterochronic gene lin-4 in C. elegans was found not to encode a protein but a small ~ 22 nt transcript complementary to the 3′ untranslated region (UTR) of lin-14 mRNA that temporally decreases LIN-14 protein levels in the first larval stage (L1) [64, 65]. Thereafter, the C. elegans heterochronic switch gene let-7 was found to encode a temporally regulated 21 nt RNA complementary to sites in the 3′UTR regions of lin-14, lin-28, lin-41, lin-42 and daf-12, thereby regulating the developmental timing of C. elegans [66]. At that time, these short RNAs were termed small temporal RNA (stRNA), which is the predecessor of the term miRNA.
MiRNAs are key players in post-transcriptional gene regulation that target mRNAs, and they have been reported in various multicellular animals and plants as well as viruses [67, 68]. In 2001, three groups published their findings in the same issue of Science, reporting the prediction of precursor structures of Drosophila melanogaster (D. melanogaster) miRNAs and human miRNAs [69], the identification of 55 C. elegans miRNAs [70] and 15 new C. elegans miRNAs through a combination of cloning and bioinformatic approaches [10].
MiRNAs in the dicot A. thaliana were first isolated in 2002, followed by the functional characterization of many key miRNA metabolism factors. For example, DICER-LIKE 1 (DCL1) and HUA ENHANCER 1 (HEN1) are required for miRNA accumulation in A. thaliana [1, 3, 18]. In the monocot rice, some new miRNAs evolved independently, based on comparison with Arabidopsis [7]. In bryophytes, the oldest land plants, miRNAs have also been identified and are considered to play important regulatory roles [5]. The wide distribution of miRNAs and diversity of species hosting this RNA regulatory mechanism suggest that miRNAs have extensive functions in eukaryotic cells.
MiRNAs were once thought to exist only in higher multicellular eukaryotes as a gene expression regulatory system to increase biocomplexity. In 2007, miRNAs were reported in lower unicellular green alga C. reinhardtii by two independent research groups using large-scale high-throughout sequencing analysis [8, 9]. They showed that miRNAs in C. reinhardtii could cut target mRNAs in vivo and in vitro and also found that the presumptive miRNA precursors were similar to those of higher plants. Further research showed that the expression of some miRNAs (or candidate miRNAs) increased or decreased during C. reinhardtii gametogenesis, indicating spatiotemporal specificity of these miRNAs. Additionally, there were other kinds of sRNAs reported, including small interfering RNAs (siRNAs) similar to trans-acting siRNAs in plants and some siRNAs derived from protein-coding genes and transposons [9]. The common features of miRNAs between unicellular algae and higher plants indicate that the complex miRNA post-transcriptional regulatory system evolved earlier than multicellular organisms and may be an ancient regulatory mechanism [8, 9].
Many new miRNAs have been characterized through stress treatment of C. reinhardtii. We predicted 85 known miRNAs and 225 novel miRNAs from sulfur-deprived C. reinhardtii; among these miRNAs, 47 were differentially expressed (mostly upregulated) and 13 were specific to the sulfur-deprived condition [71, 72], along with genome-wide long non-coding RNA were screened, identified and characterized [73]. These findings strongly suggested that sulfur deprivation significantly influences miRNA expression patterns.
Many other miRNAs in C. reinhardtii have been identified based on their association with key proteins. For example, AGO3-associated miRNAs were identified by affinity purification in C. reinhardtii, which yielded 45 unique miRNAs including 32 that were previously unknown [74]. Mutants in the DICER-LIKE 3 (DCL3) gene failed to produce miRNAs, indicating that most miRNAs are produced by DCL3-mediated cleavage in C. reinhardtii [33]. Recently, pri-miRNAs associated with the RNA-binding protein Dull slicer 16 (DUS16) were characterized in C. reinhardtii by sequencing the sRNAs of the dus16 mutant; 35 pri-miRNA genes were predicted de novo, 9 of which were novel [35]. The high degree of overlap between DUS16-dependant pri-miRNAs and DCL3-dependant pri-miRNAs indicates a cooperative relationship between DUS16 and DCL3 in C. reinhardtii pri-miRNA processing.
MiRNA biogenesis
MiRNA biogenesis involves two steps in both animals and plants. In animals, the long, capped and polyadenylated pri-miRNAs are synthesized by RNA polymerase II then cut by the RNAse III enzyme Drosha, acting with double-stranded RNA-binding (DRB) protein Pasha and Ars2 [75, 76], creating a ~ 70 nt stem loop known as a precursor miRNA (pre-miRNA). The pre-miRNA is exported from the nucleus to the cytoplasm by Exportin 5, and then cut into a miRNA/miRNA* duplex with 2 nt 3′ overhangs by Dicer, another RNAse III enzyme. The dsRBD proteins are also involved in this process, such as Loquacious (Loqs) in Drosophila and the trans-activator RNA (tar)-binding protein (TRBP) in humans [77]. One strand of the duplex is degraded, and the other single-stranded mature miRNA is loaded onto RISC, whose core component is an AGO protein [78] (Fig. 1). In plants, the Dicer homolog DCL1, assisted by the RNA-binding proteins HYPONASTIC LEAVES1 (HYL1/DRB1) and SERRATE (SE), is responsible for both steps of miRNA processing to produce the miRNA/miRNA* duplex in the nucleus. The duplex is subsequently transported to the cytoplasm by HASTY [79, 80] (Fig. 1).
There are notable differences in miRNA biogenesis between plants and animals. First, the locations of miRNA-encoding genes differ. In plants, the majority of these genes are located in intergenic regions between protein-coding genes and have independent transcription elements. In animals, most miRNA-encoding genes reside in the introns of protein-coding genes. Second, plant miRNAs are synthesized entirely within the nucleus, with the two cleavage steps performed by the RNAse III enzyme DCL1, which is endonuclear. The first cleavage of animal miRNA processing takes place in the nucleus, while the second cleavage occurs in the cytoplasm [81, 82]. Third, miRNA length differs between plants and animals. Plant miRNAs are typically 21 nt, while animal miRNAs are a little longer at 22–24 nt, which may reflect the different enzymes responsible for the first processing cleavage (DCL1 in plants and Drosha in animals) (Fig. 1). Finally, most plant miRNAs are methylated at the 3′ end to protect them from degradation after the two cleavage steps, whereas few animal miRNAs exhibit such modifications.
A recent study in C. reinhardtii found that roughly 50% of Chlamydomonas miRNA genes are embedded within the introns of protein-coding genes, which is similar to the finding in animals that approximately 30% of miRNA genes are located in the introns of protein-coding genes [33, 41, 83]. In contrast, few cases of intronic miRNA genes are known in plants; instead, plant miRNA genes typically occur in intergenic regions. Among three DCL paralogs, DCL3 of C. reinhardtii (CrDCL3) is in charge of cleaving pri-miRNAs into mature miRNAs, functioning similarly to Drosha and Dicer in animals and DCLs in plants. Interestingly, CrDCL3 lacks a PAZ domain but has a proline-rich region, which is similar to Drosha in animals (Fig. 1) [33, 84]. In unicellular C. reinhardtii, miRNA biogenesis is poorly understood but seems to be more similar to the process in metazoans than in land plants.
As with the essential partners that interact with the nucleases DCL and Drosha in multicellular organisms, a DRB protein is similarly involved in the precise processing of pri-miRNAs in C. reinhardtii. In A. thaliana, HYL1/DRB1 and DRB2 cooperate with DCL1 to ensure efficient and precise cleavage of pri-miRNAs [20, 22, 23, 85,86,87,88,89,90], with HYL1/DRB1 directing guide strand selection from the miRNA duplex [26]. To date, only one DRB protein, DUS16, has been implicated in pri-miRNA processing in C. reinhardtii. DUS16, as a component of a microprocessor complex, interacts with CrDCL3 and recognizes pri-miRNA transcripts cotranscriptionally [91], which is similar to the functions of DRB proteins involved in miRNA biogenesis in animals and land plants (Fig. 1). In our research, we have isolated and identified several other DRBs whose loss leads to reduced miRNA abundance, especially miRNA B (miR B) and miRNA C (miR C) (data not yet published).
No miRNAs appear to be conserved between Chlamydomonas and land plants. Moreover, only one miRNA is homologous between Chlamydomonas and Volvox carteri, another green alga [41, 92,93,94,95,96]. This suggests a high divergence rate for miRNAs between microalgae and land plants, and even between different green algal lineages. Thus, changes in the miRNA pathway from unicellular to multicellular organisms appear to reflect a complex and rapid evolution.
Modification of miRNAs
Methylation of miRNAs
In contrast to animal miRNAs, mature miRNAs in plants undergo 2′-O-methylation modification by HEN1 at the 3′ end, which helps to maintain their stability and helps prevent exonucleolytic degradation. HEN1 was initially identified as a floral pattering gene with multiple roles in floral organ identity specification. In Arabidopsis hen1 mutants, miRNA abundance was found to be largely reduced, while miRNA size increased, with one–five U residues added to the 3′ ends of miRNAs. In vitro, HEN1 recognizes and acts on miRNA/miRNA* duplexes but not on pre-miRNAs or single-stranded miRNA or miRNA* [18, 27,28,29, 31, 97, 98]. Further research showed that this methylation modification at the 3′ end also occurs on plant siRNAs and animal Piwi-interacting RNAs (piRNAs) [99,100,101,102,103,104].
The C. reinhardtii homolog of HEN1 is poorly characterized, and it is unknown whether Chlamydomonas miRNAs require methylation by HEN1. We have cloned the HEN1 gene of C. reinhardtii and predicted and analyzed its protein structure, which harbors a methyltransferase domain. We also observed that miRNA abundance in the Chlamydomonas hen1 mutant is generally reduced (data not yet published).
Uridylation of miRNAs
Plant miRNAs that are not methylated are subject to 3′ end uridylation by a poly(U) polymerase, resulting in subsequent degradation. In C. elegans, the 5′-to-3′ exoribonuclease XRN2 is responsible for the degradation of mature miRNAs [105]. Recent studies have associated 3′ truncation and 3′ uridylation with miRNA degradation [99, 103, 106, 107].
Nucleotidyl transferases, such as HEN1 suppressor 1 (HESO1), play a crucial role in miRNA uridylation in Arabidopsis; specifically, these enzymes may lead to the uridylation of AGO1-bound unmethylated miRNAs to trigger their degradation [108,109,110,111]. The 3′-to-5′ exoribonuclease SMALL RNA DEGRADING NUCLEASE (SDN) family is also implicated in the turnover and degradation of miRNAs in A. thaliana. A recent study showed the involvement of two members of the SDN family, SDN1 and SDN2, in miRNA 3′ truncation; the 3′ truncated miRNAs were subsequently tailed by HESO1, leading to their degradation in Arabidopsis. In addition, these SDNs were also required for the degradation of AGO10-bound miR165/6 [107, 109, 112].
In C. elegans and mammalian cells, miRNA let-7 regulates lin-28, whereas lin-28 binds to the let-7 precursor in the cytoplasm and stimulates 3′ tail uridylation by a poly(U) polymerase, resulting in precursor degradation and reduced let-7 miRNA [113,114,115,116]. Nonetheless, miRNA uridylation does not always lead to degradation. For instance, uridylation of mature miR-26 reduces its activity in terms of target regulation without affecting its abundance in mammalian cells [115].
In Chlamydomonas, miRNA degradation is not well understood, but the RNAi defective mutant Mut-68, lacking the nucleotidyltransferase MUT68, has shed some light on the degradation of miRNAs and siRNAs [117] MUT68 is involved in 3′ end uridylation of miRNAs in vivo and may mediate miRNA degradation in cooperation with RRP6, a subunit of the exosome (a 3′-to-5′ exonuclease); depleting RRP6 also increases miRNA and siRNA accumulation in vivo. MUT68 carries out the degradation of longer mRNAs generated by RISC cleavage, and it may preferentially uridylate small RNAs and adenylate RISC-cleaved transcripts. Ultimately, degradation is a quality control mechanism for the removal of dysfunctional or damaged small RNAs [118, 119], and the processes of miRNA degradation and biogenesis must be balanced for proper regulatory function in cells.
Mode of action and utilization of miRNAs
MiRNA mode of action in C. reinhardtii
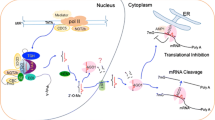
Mature miRNAs are loaded onto AGO-containing RISC, which performs cleavage and/or translation repression of target mRNAs through the complementarity of miRNAs to their target mRNAs. Plant miRNAs often exhibit almost perfect complementarity to their target mRNAs, leading to endonucleolytic slicing between positions 10 and 11 of miRNA/mRNA hybrids and subsequent degradation of the target mRNA (Fig. 2) [2, 4, 6, 39, 41, 42, 120, 121]. The complementarity of animal miRNAs to their target mRNAs is usually not as strict: only the seed sequence (positions 2–8 of the miRNA) needs to be perfectly paired, with imperfect hybridization permissible at positions 9–12 of the miRNA central region (Fig. 2) [6, 120]. Due to the loose sequence complementarity, a given animal miRNA is often predicted to have multiple target mRNAs, whereas a given plant miRNA would have far fewer targets. In terms of mode of action, animal miRNAs often induce translational repression of targets by blocking translation initiation or elongation or by deadenylation (Fig. 2), resulting in a relatively weaker modulatory effect of target mRNA and protein levels compared to cleavage [121,122,123,124,125]. Cleavage and translation repression are the common modes of action in plants and animals, respectively, exceptions abound, including miRNA-induced translation inhibition in plants and mRNA cleavage in animals [39, 126, 127]. MiRNA-mediated translation inhibition in plants also requires perfect complementarity and depends on the GW-repeat protein SUO, which is not homologous to the animal counterpart GW182 [41, 128, 129]. These findings suggest that a given miRNA may simultaneously carry out two modes of action at the same time, with one mode being dominant.
MiRNA modes of action in animals, plants and C. reinhardtii. Mature miRNAs are indicated in red, and mRNAs are in blue. The open reading frames of genes are represented with purple boxes. a In animals, mature miRNAs bind to the 3′UTR of their target mRNAs to trigger translation repression. The complementarity of animal miRNAs to their target mRNAs is imperfect: only the seed sequence (positions 2–8 of the miRNA) needs to be perfectly paired. Animal miRNAs often induce translational repression of targets by blocking translation initiation or elongation. b In plants, mature miRNAs bind to the coding region of their target mRNAs. Plant miRNAs typically exhibit near-perfect complementarity to their target mRNAs, leading to endonucleolytic slicing between positions 10 and 11 of miRNA/mRNA hybrids and subsequent degradation of the target mRNA. c In Chlamydomonas, mature miRNAs prefer to bind to the 3′UTR of their target mRNAs to trigger translation repression, similar to animal miRNAs. The complementarity of Chlamydomonas miRNAs to their target mRNAs is imperfect; perfect base pairing of the seed sequence is sufficient to induce moderate repression of target mRNAs
MiRNA regulatory mechanisms remain poorly understood in unicellular C. reinhardtii compared to multicellular eukaryotes. A mismatch tolerance assay of Chlamydomonas miRNA cre-miR1174.2 showed that complementary pairing of the 5′ end seed sequence was sufficient to trigger moderate repression of the target transcript, while pairing of the 3′ end sequence between the miRNA and its target was not significant for post-transcriptional regulation [42]. These features of Chlamydomonas miRNA target recognition differ from the near perfect complementarity required for miRNAs in higher plants but are similar to the characteristics of metazoan miRNAs (Fig. 2) [33, 130]. Thus, a given miRNA may have more than one target mRNA in Chlamydomonas, which would increase the difficulty of targeting a specific gene. MiRNA-mediated target mRNA cleavage also exists in Chlamydomonas [131].
In Chlamydomonas, there are three AGO paralogs (AGO1, AGO2 and AGO3), and AGO3 is critical for miRNA-mediated target transcript cleavage and translational repression. In contrast, AGO2 appears to have a limited contribution to gene silencing, and AGO1 is potentially uninvolved in miRNA-mediated post-transcriptional activity [34]. Another difference appears to exist between the miRNA-mediated RNA silencing mechanisms of Chlamydomonas and plants. Transcriptome analysis revealed more transcripts in the dcl3 mutant compared to wild-type, reflecting the role of DCL3 in cutting miRNA precursor structures embedded in mRNAs but not the cleavage of target mRNA [33, 94]. C. reinhardtii miRNAs bind to the 3′UTR of their target mRNAs to trigger translation repression. Moreover, this miRNA-mediated inhibition occurs at a post-initiation step of protein synthesis, which may affect the function or structural conformation of translating ribosomes [42, 132].
Artificial miRNAs and STTM in C. reinhardtii
Artificial miRNAs
Following the large-scale identification of Chlamydomonas miRNAs, artificial miRNA (amiRNA) systems were developed for knockdown in Chlamydomonas as a reverse genetics approach. For example, amiRNAs targeting the MAA7 and RBCS1/2 genes were constructed, using an endogenous Chlamydomonas miRNA precursor as the backbone that could cleave and decrease the expression of their respective target transcripts [44]. Based on the ligation of DNA oligonucleotides, a highly specific and high-throughput amiRNA system was developed for targeted gene silencing in Chlamydomonas [43]. In light of possible off-target effects, an inducible amiRNA system was developed thereafter, incorporating the NIT1 promoter, which is repressed by ammonium and activated by nitrate; heat shock factor 1 (HSF1) was the designated target, and the analysis demonstrated that HSF1 is a crucial thermotolerance regulator in Chlamydomonas [45]. In a study using amiRNA technology to target pyruvate formate lyase (PFL1), which catalyzes the conversion of pyruvate to acetyl-CoA and formate under anoxic conditions, PFL1 protein and mRNA levels were both decreased by 80–90% in two transformants [46]. An optimized one-step amiRNA precursor construction was also established for high-throughput assays, with potential utility not only in Chlamydomonas but also in other model organisms. Specifically, the Gaussia princeps luciferase (G-Luc) gene was positioned between the promoter and the amiRNA precursor to facilitate screening and detection, and the inducible promoter of the nitrate reductase gene was added to the amiRNA precursor vector construct [47].
Our previous research shows using an amiRNA strategy to knock down a photosystem II-related protein, oxygen evolving enhancer (OEE2), higher H2 production was observed in Chlamydomonas [48]. Afterwards, a blue light-inducible system was constructed to optogenetically regulate an amiRNA (amiR-D1) and, consequently, regulation of its target gene. Hydrogen production was enhanced in Chlamydomonas using this system [133, 134]. In addition, using amiRNA to inhibit the phosphoenolpyruvate carboxylase increased fatty acid production in a green microalga Chlamydomonas [133]. These particular examples have important economic potential in terms of providing sustained H2 and fatty acid productions by green algae for industrial applications, avoiding some of the inconvenience of traditional hydrogen production methods.
STTM in C. reinhardtii
In recent years, short tandem target mimic (STTM) technology has been shown to be an effective approach for blocking the functions of specific miRNAs and siRNAs in both plants and animals [49,50,51,52, 135,136,137]. However, in our research, we have observed non-specific effects when using STTM to knock down miRNAs in C. reinhardtii (data not yet published). There are several possible explanations for the differences in STTM efficacy between unicellular C. reinhardtii and multicellular plants and animals. For instance, the cellular components and the degradation mechanisms may be distinct. The optimal “spacer” length for STTM may also differ. More broadly, these differences suggest a disparate evolution rate of the miRNA pathway among plants, animals and C. reinhardtii.
Conclusions and future perspective
MiRNAs, as a class of endogenous non-coding RNAs, play an essential role in post-transcriptional gene regulation in plants and animals. Recent studies have helped to establish that miRNAs may also have an important role in unicellular microalgae. Some encouraging results have been reported, for instance, endogenous miRNA showed promising regulatory roles on H2 production in C. reinhardtii [138]. In addition, by artificial miRNAs we achieved successfully to regulate the productions of H2, fatty acid and carotenoids [133, 134, 139,140,141,142,143]. Understanding the mechanism of action of miRNAs in C. reinhardtii is of great significance to further utilize microalgae as biofuel- or bioproduct-based resources. Unlike the relatively clear mechanisms of miRNA-mediated gene regulation in plants and animals, however, the corresponding processes in C. reinhardtii remain vague. From the presently available research, we know there are important similarities and differences in the miRNA mechanisms of unicellular C. reinhardtii and multicellular organisms. MiRNA-encoding genes are found within the introns of protein-coding genes in C. reinhardtii, similar to miRNA genes in animals. Protein structure analysis of Chlamydomonas DCL3 showed that it resembles animal Drosha, which lacks a PAZ domain. Additionally, Chlamydomonas miRNAs usually bind to the 3′UTR of their targets, with seed sequence base pairing being sufficient for targeting. The mainstream academic view is that miRNA biogenesis and regulation in C. reinhardtii are more similar to that in animals than that in plants.
Research on the miRNA pathway in unicellular C. reinhardtii faces particular challenges. Compared with Arabidopsis and animals, the bigger gene size and higher GC content of key genes in the Chlamydomonas miRNA pathway make it more difficult to characterize their functions. The export protein responsible for transporting Chlamydomonas miRNAs from the nucleus to the cytoplasm is presently unknown. Is Dus16 the only DRB protein acting with CrDCL3 in Chlamydomonas miRNA biogenesis, or are there other DRBs in Chlamydomonas with different roles that affect the miRNA mode of action, as in Arabidopsis?
The localization of the core miRNA biogenesis protein CrDCL3 in C. reinhardtii is also unknown. Does miRNA processing and CrDCL3 activity occur in both the nucleus and cytoplasm, or is CrDCL3 responsible for cleavage only in the nucleus? Are mature miRNAs in C. reinhardtii methylated by HEN1? These remain open questions in Chlamydomonas (represented by gray areas in Fig. 1) to be addressed by future research. Assessing the miRNA pathway functions of C. reinhardtii proteins homologous to known miRNA-related proteins will help to improve our understanding of the evolution of the miRNA pathway in eukaryotes.
As a unicellular organism, C. reinhardtii has no tissue and organ differentiation and thus lacks apparent and easily observable phenotypes. In our studies, several loss-of-function mutants affecting critical miRNA pathway genes exhibit regular growth, whereas loss of function of these key genes in plants or animals always results in abnormal growth. Does this mean that miRNAs in unicellular C. reinhardtii are not as important as we expected? Future research regarding miRNA production and regulation in the model green alga C. reinhardtii should address this question and shed light on the evolution of miRNA mechanisms.
With the rapid improvement and reduced cost of sequencing technology, miRNAs have been identified in more and more eukaryotic organisms. Although the mechanism of action of miRNA has been only studied in model microalga C. reinhardtii at present, many other algae, such as Volvox carteri, Phaeodactylum tricornutum, Porphyridium purpureum and brown algae have been successively identified for miRNAs [95, 144,145,146]. In this regard, miRNA sequencing and annotation in other species of unicellular microalgae, combined with analysis of miRNA mode of action (cleavage of targets or translation inhibition) in future, will shed light on open questions about miRNA evolutionary turnover.
Abbreviations
- amiRNA:
-
artificial miRNA
- A. thaliana :
-
Arabidopsis thaliana
- AGO:
-
Argonaute
- C. elegans :
-
Caenorhabditis elegans
- C. reinhardtii :
-
Chlamydomonas reinhardtii
- dsRBD:
-
double-stranded RNA-binding domain
- DCL1:
-
DICER-LIKE 1
- DCL3:
-
DICER-LIKE 3
- CrDCL3:
-
DCL3 of C. reinhardtii
- D. melanogaster :
-
Drosophila melanogaster
- DUS16:
-
Dull slicer 16
- G-Luc:
-
Gaussia princeps luciferase
- HSF1:
-
heat shock factor 1
- HESO1:
-
HEN1 suppressor 1
- HEN1:
-
HUA ENHANCER 1
- HYL1/DRB1:
-
HYPONASTIC LEAVES1
- Loqs:
-
Loquacious
- miRNAs:
-
microRNAs
- miR B:
-
miRNA B
- miR C:
-
miRNA C
- OEE2:
-
oxygen evolving enhancer
- piRNAs:
-
Piwi-interacting RNAs
- pre-miRNA:
-
precursor miRNA
- pri-miRNAs:
-
primary miRNAs
- PFL1:
-
pyruvate formate lyase
- RNase III:
-
ribonuclease III
- RISC:
-
RNA-induced silencing complex
- SE:
-
SERRATE
- STTM:
-
short tandem target mimic
- siRNAs:
-
small interfering RNAs
- stRNA:
-
small temporal RNA
- SDN:
-
SMALL RNA DEGRADING NUCLEASE
- TRBP:
-
trans-activator RNA (tar)-binding protein
- UTR:
-
untranslated region
References
Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14(7):1605–19.
Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297(5589):2053–6.
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16(13):1616–26.
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110(4):513–20.
Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43(6):837–48.
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20.
Sunkar R, Girke T, Jain PK, Zhu JK. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17(5):1397–411.
Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447(7148):1126–9.
Zhao T, Li GL, Mi SJ, Li S, Hannon GJ, Wang XJ, et al. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21(10):1190–203.
Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–4.
Han JJ, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–27.
Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–5.
Yeom KH, Lee Y, Han JJ, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34(16):4622–9.
Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6.
Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–8.
Chendrimada TP, Finn KJ, Ji XJ, Baillat D, Gregory RI, Liebhaber SA, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447(7146):823.
Wynant N, Santos D, Broeck JV. The evolution of animal Argonautes: evidence for the absence of antiviral AGO Argonautes in vertebrates. Sci Rep. 2017;7:9230.
Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol (CB). 2002;12(17):1484–95.
Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7(11):487–91.
Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 2004;101(4):1093–8.
Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. P Natl Acad Sci USA. 2004;101(34):12753–8.
Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol (CB). 2004;14(4):346–51.
Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12(2):206–12.
Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 2006;7(10):1052–8.
Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47(6):841–50.
Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA. 2009;15(12):2219–35.
Boutet S, Vazquez F, Liu J, Beclin C, Fagard M, Gratias A, et al. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr Biol (CB). 2003;13(10):843–8.
Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol (CB). 2005;15(16):1501–7.
Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307(5711):932–5.
Ren G, Xie M, Zhang S, Vinovskis C, Chen X, Yu B. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc Natl Acad Sci USA. 2014;111(17):6365–70.
Zhao Y, Chen X. Noncoding RNAs and DNA methylation in plants. Natl Sci Rev. 2014;1(2):219–29.
Baranauske S, Mickute M, Plotnikova A, Finke A, Venclovas C, Klimasauskas S, et al. Functional mapping of the plant small RNA methyltransferase: hEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins. Nucleic Acids Res. 2015;43(5):2802–12.
Valli AA, Santos BACM, Hnatova S, Bassett AR, Molnar A, Chung BY, et al. Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by Dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs. Genome Res. 2016;26(4):519–29.
Yamasaki T, Kim EJ, Cerutti H, Ohama T. Argonaute3 is a key player in miRNA-mediated target cleavage and translational repression in Chlamydomonas. Plant J. 2016;85(2):258–68.
Yamasaki T, Cerutti H. Cooperative processing of primary miRNAs by DUS16 and DCL3 in the unicellular green alga Chlamydomonas reinhardtii. Commun Integr Biol. 2017;10(1):e1280208.
Vaucheret H. Plant argonautes. Trends Plant Sci. 2008;13(7):350–8.
Mallory A, Vaucheret H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010;22(12):3879–89.
Zhang Z, Hu F, Sung MW, Shu C, Castillo-Gonzalez C, Koiwa H, et al. RISC-interacting clearing 3′-5′ exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. eLife. 2017;6:e24466.
Liu Q, Wang F, Axtell MJ. Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell. 2014;26(2):741–53.
Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21(3):452–60.
Moran Y, Agron M, Praher D, Technau U. The evolutionary origin of plant and animal microRNAs. Nat Ecol Evol. 2017;1(3):27.
Yamasaki T, Voshall A, Kim EJ, Moriyama E, Cerutti H, Ohama T. Complementarity to an miRNA seed region is sufficient to induce moderate repression of a target transcript in the unicellular green alga Chlamydomonas reinhardtii. Plant J. 2013;76(6):1045–56.
Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, et al. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009;58(1):165–74.
Zhao T, Wang W, Bai X, Qi YJ. Gene silencing by artificial microRNAs in Chlamydomonas. Plant J. 2009;58(1):157–64.
Schmollinger S, Strenkert D, Schroda M. An inducible artificial microRNA system for Chlamydomonas reinhardtii confirms a key role for heat shock factor 1 in regulating thermotolerance. Curr Genet. 2010;56(4):383–9.
Burgess SJ, Tredwell G, Molnar A, Bundy JG, Nixon PJ. Artificial microRNA-mediated knockdown of pyruvate formate lyase (PFL1) provides evidence for an active 3-hydroxybutyrate production pathway in the green alga Chlamydomonas reinhardtii. J Biotechnol. 2012;162(1):57–66.
Hu J, Deng X, Shao N, Wang G, Huang K. Rapid construction and screening of artificial microRNA systems in Chlamydomonas reinhardtii. Plant J. 2014;79(6):1052–64.
Li H, Zhang L, Shu L, Zhuang X, Liu Y, Chen J, et al. Sustainable photosynthetic H2-production mediated by artificial miRNA silencing of OEE2 gene in green alga Chlamydomonas reinhardtii. Int J Hydrogen Energy. 2015;40(16):5609–16.
Tang GL, Yan J, Gu YY, Qiao MM, Fan RW, Mao YP, et al. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods. 2012;58(2):118–25.
Yan J, Gu YY, Jia XY, Kang WJ, Pan SJ, Tang XQ, et al. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell. 2012;24(2):415–27.
Tang GL, Tang XQ. Short tandem target mimic: a long journey to the engineered molecular landmine for selective destruction/blockage of MicroRNAs in plants and animals. J Genet Genomics. 2013;40(6):291–6.
Reichel M, Li YJ, Li JY, Millar AA. Inhibiting plant microRNA activity: molecular SPONGEs, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol J. 2015;13(7):915–26.
Zhang H, Zhang JS, Yan J, Gou F, Mao YF, Tang GL, et al. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. P Natl Acad Sci USA. 2017;114(20):5277–82.
Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17(1):170–80.
Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18(10):1187–97.
Zhang XM, Zhao HW, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, et al. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a golgi-localized SNARE gene, MEMB12. Mol Cell. 2011;42(3):356–66.
Qi YJ, He XY, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443(7114):1008–12.
Hunter C, Wu G, Sun H, Poethig RS. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Dev Biol. 2003;259(2):521–2.
Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, et al. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426(6966):579–84.
van den Berg A, Mols J, Han JH. RISC-target interaction: cleavage and translational suppression. BBA-Gene Regul Mech. 2008;1779(11):668–77.
Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–60.
Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16(6):720–8.
Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. P Natl Acad Sci USA. 2003;100(17):9779–84.
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity To lin-14. Cell. 1993;75(5):843–54.
Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern-formation in C. elegans. Cell. 1993;75(5):855–62.
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–6.
Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–2.
Bauman Y, Nachmani D, Vitenshtein A, Tsukerman P, Drayman N, Stern-Ginossar N, et al. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe. 2011;9(2):93–102.
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8.
Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–62.
Longfei Shu ZH. Small silencing RNAs in Chlamydomonas reinhardtii. Minerva Biotecnol. 2010;22(1):9.
Shu LF, Hu ZL. Characterization and differential expression of microRNAs elicited by sulfur deprivation in Chlamydomonas reinhardtii. BMC Genomics. 2012;13:108.
Li H, Wang YT, Chen MR, Xiao P, Hu CX, Zeng ZY, et al. Genome-wide long non-coding RNA screening, identification and characterization in a model microorganism Chlamydomonas reinhardtii. Sci Rep. 2016;6:34109.
Voshall A, Kim EJ, Ma X, Moriyama EN, Cerutti H. Identification of AGO3-associated miRNAs and computational prediction of their targets in the green alga Chlamydomonas reinhardtii. Genetics. 2015;200(1):105–21.
Wilson MD, Wang D, Wagner R, Breyssens H, Gertsenstein M, Lobe C, et al. ARS2 is a conserved eukaryotic gene essential for early mammalian development. Mol Cell Biol. 2008;28(5):1503–14.
Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, et al. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138(2):340–51.
Du TT, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132(21):4645–52.
Du T, Zamore PD. Beginning to understand microRNA function. Cell Res. 2007;17(8):661–3.
Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development. 2003;130(8):1493–504.
Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA. 2005;102(10):3691–6.
Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106(1):23–34.
Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15(20):2654–9.
Axtell MJ, Westholm JO, Lai EC. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12(4):221.
Cui J, You CJ, Chen XM. The evolution of microRNAs in plants. Curr Opin Plant Biol. 2017;35:61–7.
Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA. 2008;105(29):9970–5.
Pouch-Pelissier MN, Pelissier T, Elmayan T, Vaucheret H, Boko D, Jantsch MF, et al. SINE RNA induces severe developmental defects in Arabidopsis thaliana and interacts with HYL1 (DRB1), a key member of the DCL1 complex. PLoS Genet. 2008;4(6):e1000096.
Eamens AL, Kim KW, Curtin SJ, Waterhouse PM. DRB2 is required for microRNA biogenesis in Arabidopsis thaliana. PLoS ONE. 2012;7(4):e35933.
Reis RS, Hart-Smith G, Eamens AL, Wilkins MR, Waterhouse PM. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nat Plants. 2015;1:14027.
Reis RS, Hart-Smith G, Eamens AL, Wilkins MR, Waterhouse PM. MicroRNA regulatory mechanisms play different roles in Arabidopsis. J Proteome Res. 2015;14(11):4743–51.
Eamens AL, Wook Kim K, Waterhouse PM. DRB2, DRB3 and DRB5 function in a non-canonical microRNA pathway in Arabidopsis thaliana. Plant Signal Behav. 2012;7(10):1224–9.
Yamasaki T, Onishi M, Kim EJ, Cerutti H, Ohama T. RNA-binding protein DUS16 plays an essential role in primary miRNA processing in the unicellular alga Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2016;113(38):10720–5.
Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of MIRNA genes. Plant Cell. 2011;23(2):431–42.
Nozawa M, Miura S, Nei M. Origins and evolution of microRNA genes in plant species. Genome Biol Evol. 2012;4(3):230–9.
Tarver JE, Donoghue PCJ, Peterson KJ. Do miRNAs have a deep evolutionary history? Bioessays. 2012;34(10):857–66.
Li JR, Wu Y, Qi YJ. microRNAs in a multicellular green alga Volvox carteri. Sci China Life Sci. 2014;57(1):36–45.
Alaba S, Piszczalka P, Pietrykowska H, Pacak AM, Sierocka I, Nuc PW, et al. The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants. New Phytol. 2015;206(1):352–67.
Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell. 2010;143(5):703–9.
Huang Y, Ji LJ, Huang QC, Vassylyev DG, Chen XM, Ma JB. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature. 2009;461(7265):823.
Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng ZP, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328(5985):1534–9.
Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol (CB). 2007;17(14):1265–72.
Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, et al. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 2010;29(21):3688–700.
Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2’-O-methylated at their 3’ termini. Nat Struct Mol Biol. 2007;14(4):347–8.
Li JJ, Yang ZY, Yu B, Liu J, Chen XM. Methylation protects miRNAs and siRNAs from a 3 ‘-end uridylation activity in Arabildopsis. Curr Biol. 2005;15(16):1501–7.
Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2’-O-methylation of PIWI-interacting RNAs at their 3’ ends. Genes Dev. 2007;21(13):1603–8.
Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461(7263):546.
Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJT, Bagijn MP, Sapetschnig A, et al. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 2012;8(7):1002702.
Yu Y, Ji L, Le BH, Zhai J, Chen J, Luscher E, et al. ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 2017;15(2):e2001272.
Ren G, Chen X, Yu B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr Biol (CB). 2012;22(8):695–700.
Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, et al. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol (CB). 2012;22(8):689–94.
Tu B, Liu L, Xu C, Zhai JX, Li SB, Lopez MA, et al. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in MicroRNA turnover in Arabidopsis. PLoS Genet. 2015;11(4):e1005119.
Wang XY, Zhang SX, Dou YC, Zhang C, Chen XM, Yu B, et al. Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3’ tailing of small RNAs in Arabidopsis. PLoS Genet. 2015;11(4):e1005091.
Ramachandran V, Chen XM. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321(5895):1490–2.
Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16(10):1021.
Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through Pre-MicroRNA uridylation. Cell. 2009;138(4):696–708.
Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu SN, Ivanov AR, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009;11(9):1157.
Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, et al. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16(10):1016.
Ibrahim F, Rohr J, Jeong WJ, Hesson J, Cerutti H. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science. 2006;314(5807):1893.
Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, et al. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. P Natl Acad Sci USA. 2010;107(8):3906–11.
Cerutti H, Ibrahim F. Turnover of mature miRNAs and siRNAs in plants and algae. Adv Exp Med Biol. 2011;700:124–39.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33.
Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–87.
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8(4):517–27.
Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71.
Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14.
Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63.
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320(5880):1185–90.
Iwakawa H, Tomari Y. Molecular insights into microRNA-mediated translational repression in plants. Mol Cell. 2013;52(4):591–601.
Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA. 2009;15(8):1433–42.
Yang L, Wu G, Poethig RS. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. P Natl Acad Sci USA. 2012;109(1):315–20.
Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17(1):49–63.
Chung BYW, Deery MJ, Groen AJ, Howard J, Baulcombe DC. Endogenous miRNA in the green alga Chlamydomonas regulates gene expression through CDS-targeting. Nat Plants. 2017;3(10):787–94.
Ma XR, Kim EJ, Kook I, Ma FR, Voshall A, Moriyama E, et al. Small interfering RNA-mediated translation repression alters ribosome sensitivity to inhibition by cycloheximide in Chlamydomonas reinhardtii. Plant Cell. 2013;25(3):985–98.
Wang YT, Jiang XQ, Hu CX, Sun T, Zeng ZY, Cai XQ, et al. Optogenetic regulation of artificial microRNA improves H-2 production in green alga Chlamydomonas reinhardti. Biotechnol Biofuels. 2017;10:257.
Li H, Liu YM, Wang YT, Chen MR, Zhuang XS, Wang CG, et al. Improved photobio-H2 production regulated by artificial miRNA targeting psbA in green microalga Chlamydomonas reinhardtii. Biotechnol Biofuels. 2018;11:36.
Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–6.
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39(8):1033–7.
Gu ZH, Huang CJ, Li FF, Zhou XP. A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol J. 2014;12(5):638–49.
Wang YT, Zhuang XS, Chen MR, Zeng ZY, Cai XQ, Li H, et al. An endogenous microRNA (miRNA1166.1) can regulate photobio-H2 production in eukaryotic green alga Chlamydomonas reinhardtii. Biotechnol Biofuels. 2018;11:126.
Wang CG, Chen X, Li H, Wang JX, Hu ZL. Artificial miRNA inhibition of phosphoenolpyruvate carboxylase increases fatty acid production in a green microalga Chlamydomonas reinhardtii. Biotechnol Biofuels. 2017;10:91.
Gao CF, Zhai Y, Ding Y, Wu QY. Application of sweet sorghum for biodiesel production by heterotrophic microalga Chlorella protothecoides. Appl Energ. 2010;87(3):756–61.
Ng IS, Tan SI, Kao PH, Chang YK, Chang JS. Recent developments on genetic engineering of microalgae for biofuels and bio-based chemicals. Biotechnol J. 2017;12(10):1600644.
Kong F, Romero IT, Warakanont J, Li-Beisson Y. Lipid catabolism in microalgae. New Phytol. 2018;218(4):1340–8.
Liang MH, Zhu J, Jiang JG. High-value bioproducts from microalgae: strategies and progress. Crit Rev Food Sci Nutr. 2018;1–53.
Huang AY, He LW, Wang GC. Identification and characterization of microRNAs from Phaeodactylum tricornutum by high-throughput sequencing and bioinformatics analysis. BMC Genomics. 2011;12:337.
Gao F, Nan FR, Feng J, Lv JP, Liu Q, Xie SL. Identification of conserved and novel microRNAs in Porphyridium purpureum via deep sequencing and bioinformatics. BMC Genomics. 2016;17:612.
Cock JM, Liu FL, Duan DL, Bourdareau S, Lipinska AP, Coelho SM, et al. Rapid evolution of microRNA loci in the brown algae. Genome Biol Evol. 2017;9(3):740–9.
Authors’ contributions
SLL wrote the manuscript. TS revised the figures. HL and ZLH guided and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank Prof. Chen Xuemei for advice on text structure.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (31800300, 31470431 and 31870343), Guangdong Natural Science Foundation (2014A030308017, 2016A030313052, 2018A030310445 and 2018A030313507), Project of DEGP (2015KTSCX125), Shenzhen Grant Plan for Science & Technology (Shenzhen basic research projects, Subject layout project, CKCY2016042710211071, JCYJ20170302144605664), Demonstration Project for Marine Economic Development in Shenzhen, China’s State Oceanic Administration, Natural Science Foundation of SZU (Grant No. 2017050) and China Postdoctoral Science Foundation (2016M602507).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lou, S., Sun, T., Li, H. et al. Mechanisms of microRNA-mediated gene regulation in unicellular model alga Chlamydomonas reinhardtii. Biotechnol Biofuels 11, 244 (2018). https://doi.org/10.1186/s13068-018-1249-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-018-1249-y