Abstract
Background
The rising incidence of renal cell carcinomas (RCC) constitutes a significant challenge owing to risk of overtreatment. Because aberrant microRNA (miR) promoter methylation contributes to cancer development, we investigated whether altered miR-30a-5p expression associates with DNA promoter methylation and evaluated the usefulness as clear cell RCC (ccRCC) diagnostic and prognostic markers.
Methods
Genome-wide methylome and RNA sequencing data from a set of ccRCC and normal tissue samples from The Cancer Genome Atlas (TCGA) database were integrated to identify candidate CpG loci involved in cancer onset. MiR-30a-5p expression and promoter methylation were quantitatively assessed by PCR in a tissue set (Cohort #1) and urine sets (Cohorts #2 and 3) from IPOPorto and Homburg University Hospital. Non-parametric tests were used for comparing continuous variables. MiR-30a-5p promoter methylation (miR-30a-5pme) performance as diagnostic (receiver operator characteristics [ROC] - validity estimates) and prognostic [metastasis-free (MFS) and disease-specific survival (DSS)] biomarker was further validated in urine samples from ccRCC patients by Kaplan Meier curves (with log rank) and both univariable and multivariable analysis.
Results
Two significant hypermethylated CpG loci in TCGA ccRCC samples, correlating with miR-30a-5p transcriptional downregulation, were disclosed. MiR-30a-5pme in ccRCC tissues was confirmed in an independent patient’s cohort of IPOPorto and associated with shorter time to relapse. In urine samples, miR-30a-5pme levels identified cancer both in testing and validation cohorts, with 83% sensitivity/53% specificity and 63% sensitivity/67% specificity, respectively. Moreover, higher miR-30a-5pme levels independently predicted metastatic dissemination and survival.
Conclusion
To the best of our knowledge, this is the first study validating the diagnostic and prognostic potential of miR-30a-5pme for ccRCC in urine samples, providing new insights for its clinical usefulness as non-invasive cancer biomarker.
Similar content being viewed by others
Background
Renal cell carcinoma (RCC) is the third most prevalent urologic malignancy [1], twice more common in men than in women [2] disclosing rising incidence (2–4% per year) worldwide. RCC accounts for 2–3% of all malignant tumors in adults, displaying the highest mortality rate among urinary tract cancers [1, 3]. Notwithstanding, renal cell tumors are morphologically and genetically heterogeneous [4]. The three main subtypes of RCC - clear cell RCC (ccRCC), papillary RCC and chromophobe RCC [1] - have distinct clinical behaviors, which should be considered for adequate patient management [4].
CcRCC is simultaneously the most common and one of the most aggressive RCC subtypes, being prone to local invasion, metastization and death [2, 5]. It comprises 70–75% of all RCC cases and is characterized by several distinct genetic and epigenetic alterations [6]. About 25% of ccRCC patients present distant metastases at time of diagnosis, and in 20–50%, metastatic disease develops few years after diagnosis and surgical treatment of the primary tumor [3, 5, 7, 8]. Furthermore, ccRCC is extremely resistant to radiation and to conventional chemotherapy [3]. Therefore, biomarkers allowing for earlier diagnosis and accurate prognostication are required, improving current treatment and follow-up strategies [9]. Indeed, metastatic dissemination is the most important prognostic factor in ccRCC [10], highlighting the importance of accurately identifying patients at high risk of disease progression. Moreover, the identification of molecular biomarkers that might indicate risk of disease progression (recurrence, metastization) at the time of diagnosis might improve clinical management [3, 7] effectively contributing to implementation of Precision Medicine [10].
MicroRNAs (miRs) are small non-coding RNAs, 18–25 nucleotides long, that repress specific genes’ expression by targeting its 3′ untranslated region [11,12,13]. MiRs’ deregulation has been shown to participate in tumorigenesis, affecting differentiation, invasion, migration and apoptosis [10, 14] and has been implicated in urological tumors [15]. Several studies have associated microRNAs (miRs) deregulation with ccRCC clinicopathological features, suggesting a role in tumor initiation and progression [5, 7, 16, 17]. MiR-30a-5p, an intergenic miR (chromosome 6, 71,403,551–71,403,621 [− strand]), was suggested to play a role in cellular differentiation and development [18], but its precise role remains largely unknown [19, 20]. In ccRCC, an onco-suppressor function was proposed for miR-30a-5p, since its downregulation was associated with metastasis development [5, 9]. Moreover, miR-30a-5p was found to inhibit autophagy, by targeting BECN1, the gene encoding for beclin-1, a key protein for autophagosome formation [3]. In addition, miR-30a-5p was shown to decrease tumor microvessel density, by targeting endothelial DLL4, which is enrolled in tumor angiogenesis [5]. However, the mechanism underlying miR-30a-5p downregulation in ccRCC remains elusive [21]. Similarly to protein coding genes, miRs’ downregulation might be associated with aberrant promoter methylation, a common feature of urological tumors [22,23,24,25]. Thus, we sought to investigate for the first time, whether miR-30a-5p expression is regulated by promoter hypermethylation in ccRCC and evaluate its value as diagnostic and prognostic biomarker, both in tissue and urine samples.
Materials and methods
Patients and sample collection
Independent patient cohorts, two retrospective and one prospective, were selected for this study. Cohort #1 comprises 235 ccRCC patients, consecutively diagnosed and treated with nephrectomy, at Portuguese Oncology Institute of Porto (IPO Porto) between 2000 and 2017. For control purposes, normal kidney tissue from 25 patients subjected to nephrectomy due to upper urinary tract urothelial carcinoma was obtained. Tissue samples from primary tumors and normal kidney were collected immediately after surgery and promptly frozen at − 80 °C. Frozen tissue samples were cut in a cryostat and tumor cell content over 70% was confirmed in two hematoxylin and eosin stained slides taken before and after frozen section collection for nucleic acid extraction. A second cohort composed of 53 ccRCC patients, primarily diagnosed from 2007 to 2013 at IPO Porto, voluntarily provided 50 mL of voided urine samples (Cohort #2 - Testing). For control purposes, urine samples were collected from 57 healthy donors at IPO Porto (2009 to 2010). After collection, urine samples were centrifuged at 4000 rpm for 20 min at 4 °C and washed in PBS 1x. Lastly, pellets were frozen at − 80 °C. A third cohort (Cohort #3 – Validation) comprised 171 ccRCC patients, primarily diagnosed from 2015 to 2018 at Homburg University Hospital (Germany) provided, after informed consent, voided urine samples. For control purposes, urine samples were collected from 85 healthy donors at IPO Porto (2015–2017). After collection, 4 mL of whole urine was transferred into a tube and frozen at − 80 °C, until further usage.
Relevant clinical data was retrieved from clinical charts (Table 1). All procedures performed in studies involving human participants were performed in accordance with the ethical standards of the institutional ethics committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all participants, according institutional regulations. This study was approved by the Institutional Review Board (Comissão de Ética para a Saúde) of IPO Porto, Portugal (CES-518/2010) and Jena University Hospital IRB.
TCGA data analysis in ccRCC patients
Data on miR-30a-5p expression and methylation from ccRCC tumors and matched normal tissue samples was retrieved from The Cancer Genome Atlas (TCGA) database. MicroRNA-30a-5p expression data from samples hybridized by the University of North Carolina, Lineberger Comprehensive Cancer Center, using Illumina HiSeq 2000 Sequencing system, were downloaded from data matrix including 516 ccRCC samples (http://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp). DNA methylation data from miR-30a locus was evaluated using Illumina Infinium Human DNA Methylation 450 array and includes the methylation levels of 319 ccRCC samples. The provided value was pre-processed and normalized according to “level 3” specifications of TCGA (TCGA FPKM-UQ value; see http://cancergenome.nih.gov/ for details). This data is available for download through the NCI GDC data portal (https://portal.gdc.cancer.gov/).
DNA extraction, bisulfite modification, pre-amplification and quantitative methylation-specific PCR
DNA was extracted from all clinical samples using phenol-chloroform method. Bisulfite modification was performed using EZ DNA Methylation-Gold™ Kit (Zymo Research, Orange, CA, USA), that integrates DNA denaturation and bisulfite conversion processes into one-step, according to recommended protocol. For urine samples from cohort #3, a pre-amplification step was performed prior to the quantitative methylation-specific PCR. SsoAdvanced™ PreAmp Supermix (Bio-Rad Laboratories Inc., Hercules, CA, USA) was used, following manufacturer recommendations. In brief, 8 μL of DNA template was added to 12 μL of nuclease-free water, 5 μL of Preamplification assay pool and, 25 μL of SsoAdvanced PreAmp Supermix (2x) and pre-amplified for 12 cycles.
Quantitative Methylation-specific PCR (qMSP) assays were carried out in triplicates using Xpert Fast SYBR (Grisp, Porto, Portugal), according to recommended protocol. Sequence-specific primers used in this study were designed to include the two CpGs tested in TCGA database and synthesized by Sigma Aldrich (Sigma-Aldrich, St. Louis, MO, USA) (Supplementary Table 2). Furthermore, the primer’s coverage sites within the methylated gene are available in Supplementary Table 3. For each sample, miR-30a-5pme status was normalized to the endogenous control β-Actin.
RNA extraction
Samples were suspended in TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) and chloroform (Merk Milipore, Burlington, MA, USA) was added after cells were lysed. RNA concentrations and purity ratios were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). RNA samples were stored at − 80 °C until further usage.
MicroRNAs expression assay
Reverse transcription (RT) was performed using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) according to manufacturer’s instructions. Quantitative Real-Time PCR (RT-qPCR) was performed in triplicates using TaqMan Small RNA Assays for miR-30a-5p (Assay ID 000417, Thermo Fisher Scientific, Waltham, MA, USA) and Xpert Fast Probe (Grisp, Porto, Portugal), according to recommended protocol. For each sample, miR expression was normalized to endogenous control RNU48 (Assay ID: 001006, Thermo Fisher Scientific, Waltham, MA, USA).
Statistical analysis
Differences in methylation and expression levels and relationships between clinical variables were assessed using Kruskal-Wallis and Mann-Whitney U non-parametric tests for multiple groups (more than two) and pairwise comparisons, respectively. In multiple comparisons, Bonferroni’s correction was applied for pairwise comparisons, dividing the original P-value by the number of groups. P-values were considered statistically significant if inferior to 0.05 for comparisons between two groups.
For miR-30a-5pme, receiver operator characteristics (ROC) curves were constructed by plotting the true positive (sensitivity) against the false-positive (1-specificity) rate, and area under the curve (AUC) was calculated. Specificity, sensitivity, and accuracy were determined. For this, the empirical cut-off obtained by ROC curve analysis [sensitivity + (1-specificity)] was established. This cut-off value combines the maximum sensitivity and specificity, ensuring perfect categorization of the samples as positive and negative for methylation test.
Disease-free survival, disease-specific survival, and metastasis-free survival curves (Kaplan-Meier with log rank test) were constructed considering clinicopathological variables (stage and nuclear grade) and categorized miR-30a-5pme or expression status. A Cox-regression model (multivariable model) was computed considering all significant clinical variables, to assess the relative contribution of each variable to the follow-up status.
Statistical analysis was performed using SPSS 25.0 for Windows (SPSS Inc., Chicago, IL, USA) and graphs were built using GraphPad Prism 6.0 software for Windows (GraphPad Software, San Diego, CA, USA).
Results
Analysis of miR-30a-5pme and expression in ccRCC patients from the Cancer genome atlas
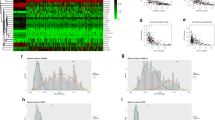
The analysis of miR-30ame locus in a cohort of ccRCC patients/samples from The Cancer Genome Atlas (TCGA) database identified two CpG sites with significantly higher methylation index (β-values) in tumor samples compared to normal controls [(P < 0.0001) (Fig. 1 (A)]. Conversely, miR-30a-5p expression levels were significantly downregulated in ccRCC when compared to non-cancerous tissue, both patient-unmatched and -matched [(P < 0·0001) (Fig. 1 (B)]. Furthermore, a significant inverse correlation between miR-30a-5p expression and promoter methylation was found [(P < 0.0001) (Fig. 1 (C)]. In univariable analysis, both miR-30a-5p expression and promoter methylation independently predicted worse overall survival (OS) and shorter recurrence-free survival (RFS). Similarly, both molecular variables retained their predictive value for OS, after adjusting for gender, age and stage, in multivariable analysis. Of note, miR-30a-5pme also remained an independent predictor of shorter RFS in this model, as well as when tumor grade was included (Supplementary Table 1).
TCGA in silico analysis. a DNA methylation levels (β-Values) for each probe in specific miR loci, comparing normal and ccRCC samples (TCGA Illumina Infinium Human DNA Methylation 450 array), both in the whole cohort (n = 319 ccRCC) and in matched tumor/normal tissue (n = 160); b TCGA RNA-seq data for miR-30a-5p expression in ccRCC samples compared to normal samples, both patient-unmatched (n = 319 ccRCC) and -matched tumor/normal tissue (n = 71); c Correlation between miR-30a-5p expression and methylation levels for each probe using TCGA dataset for ccRCC tumor samples
MiR-30-5p expression and methylation status in tissues from ccRCC patients (cohort #1)
Both promoter methylation and expression levels were assessed in an independent set of fresh-frozen tissue samples from IPO-Porto (Cohort #1). Corroborating the results from TCGA dataset, miR-30a-5pme levels were significantly higher in ccRCC, compared to normal renal tissues (RNT) [(P = 0.0086) (Fig. 2 (A)], whereas miR-30a-5p expression levels were significantly lower in ccRCC samples [(P < 0.0001) (Fig. 2 (B)]. Nonetheless, no significant inverse correlation was found between miR’s promoter methylation and expression, in this IPO Porto cohort.
MiR-30a-5p promoter methylation levels, miR-30a-5p expression levels, and prognostic value in Cohort #1. a Scatter plots representing relative miR-30a-5pme and expression levels between RNT (n = 25) and ccRCC (n = 226) samples (Mann–Whitney U test); b Disease-free and c disease-specific Kaplan-Meier survival curves based on miR-30a-5pme and expression levels (Log-rank test)
Furthermore, lower miR-30a-5p expression levels significantly associated with synchronous metastatic dissemination (P = 0.0447), recurrence (P = 0.0078) and higher nuclear grade (P = 0.0424) (Supplementary Figure 1). Conversely, no significant association was found between miR-30a-5pme levels and classical clinical indexes, namely synchronous metastatic dissemination (p = 0.545), recurrence (p = 0.128), Führman Grade (p = 0.165), stage (p = 0.816), tumor size (p = 0.161) and age (p = 0.305).
Assessment of miR-30a-5pme and expression levels as prognostic marker for ccRCC
The prognostic value of miR-30a-5p expression and promoter methylation levels were also tested. The median follow-up time of IPO Porto Cohort #1 was 61 months, range (0–194) (Table 1). For disease-free survival (DFS) analysis, seven patients were excluded, five owing to the presence of metastasis at diagnosis and two because they never accomplished disease remission. In univariable analysis, advanced pathological stage and higher nuclear grade associated with DFS [P < 0.0001 and P = 0.015, respectively; Kaplan-Meier curves shown in Supplementary Figure 2 (A) and hazard ratio from COX regression shown in Supplementary Table 4] and disease-specific survival (DSS) [P < 0·0001 and P = 0.003, respectively; Kaplan-Meier curves shown in Supplementary Figure 2 (B) and hazard ratio from COX regression shown in Supplementary Table 4], as expected. Importantly, lower miR-30a-5p expression levels (below 50th percentile) and increased miR-30a-5pme levels (over 20th percentile) significantly associated with shorter time to relapse [P = 0.030 and P = 0.035, respectively; (Fig. 2 (B)] and shorter DSS [P = 0.040 and P = 0.012, respectively; Fig. 2 (C)]. In multivariable analysis, only pathological stage, nuclear grade and miR-30a-5pme levels (20th percentile) retained independent prognostic value for DSS (Table 2; Supplementary Table 4 and 5).
MiR-30a-5pme levels in urine sediments (cohort #2 – testing set)
MiR-30a-5pme was then tested in an independent set of urine sediment samples from a retrospective cohort of patients from IPO Porto (Cohort #2 - Testing). Significantly higher miR-30a-5pme levels were found in urines from ccRCC patients compared to asymptomatic controls (AC) [(P = 0.0008) (Fig. 3 (A)]. Moreover, miR-30a-5pme levels identified malignancy with 83% sensitivity and 53% specificity, providing an overall accuracy of 67% [AUC of 0.6837, (P = 0.0009) (Fig. 3 (A)] (Table 3).
Importantly, patients that presented advanced pathological stage at diagnosis [(P = 0.0073) (Supplementary Figure 3(A)], patients who recurred (P = 0.048) and those that developed metastasis during follow up [(P = 0.0200) (Fig. 3 (B)] disclosed higher miR-30a-5pme levels. However, no further associations were found between miR-30a-5pme levels and other variables, namely age and tumor size (P = 0.294 and P = 0.224, respectively).
Remarkably, urine miR-30a-5pme levels discriminated patients with metastasis (both synchronous and metachronous) from those without metastatic disease with 80% sensitivity, 71% specificity and 73% accuracy [AUC of 0.7684, (P = 0.0025) (Fig. 3 (B)] (Table 3).
The median follow-up for cohort #2 was 58 months, range (2–91) (Table 1). Five patients with metastatic disease at diagnosis were excluded for metastasis-free survival analysis. Worse DSS and shorter metastasis-free survival (MFS) was observed in patients with advanced pathological stage and higher nuclear grade [P < 0.0001 and P = 0.002, respectively; Supplementary Figure 3 (B)] and [P < 0.0001 and P = 0.032, respectively; Supplementary Figure 3 (C)]. Remarkably, higher miR-30a-5pme (above 70th percentile) also associated with shorter MFS and worse DSS [P = 0.003 and P = 0.001, respectively; Fig. 4 (A) and (B)].
In univariable Cox-regression analysis, higher miR-30a-5pmelevels (70th percentile) and nuclear grade predicted shorter MFS and DSS (Supplementary Tables 6 and 7). However, in multivariable analysis, these parameters only depicted independent prognostic value for DSS (Table 4).
MiR-30a-5pme levels in urine supernatants (cohort #3 – validation set)
Considering the results from Cohort #2 - Testing, we further assessed the detection performance of miR-30a-5pme levels in a larger, independent set of urine from a prospective cohort of 171 ccRCC patients and 85 AC. Paralleling the previous observations, miR-30a-5pme levels were significantly higher in ccRCC patients, comparing to AC (P < 0.0001) (Fig. 5). Furthermore, miR-30a-5pme levels identified ccRCC with 63% sensitivity, 67% specificity and 63% accuracy [AUC of 0.6702, (P < 0.0001) (Fig. 5)] (Table 5). Despite miR-30a-5pme levels were lower in ccRCC stages I & II comparatively to stages III & IV, the difference did not reach statistical significance. Moreover, miR-30a-5pme did not significantly associate with either age or tumor size (p = 0.280 and p = 0.460, respectively.
Discussion
Over the last decade, the frequency of incidentally detected RCC has significantly increased, mostly due to the widespread use of imaging techniques. CcRCC, the most prevalent RCC, carries worse prognosis than other common RCC subtypes, as approximately 20–40% of cases develop distant metastases [7], which are the main cause of RCC-related mortality, the highest among urologic cancers [26, 27]. Thus, biomarkers capable of accurately identifying ccRCC cases prone to metastasize, mostly among early stage tumors at diagnosis, would be a major clinical breakthrough. MiR-30a-5p expression downregulation has been reported in ccRCC, associated with metastatic disease and adverse prognosis. This being said, we aimed to determine whether (a) miR-30a-5p expression silencing was due to aberrant promoter methylation and (b) miR-30a-5pme levels might not only accurately detect ccRCC in tissue and urine samples, but also identify patients at increased risk to develop metastatic disease independently of standard clinicopathological parameters.
Firstly, TCGA dataset was surveyed and two CpG loci at miR-30a promoter were identified as putative regulators of its expression in ccRCC. Further, miR-30a-5pme inversely correlated with miR-30a-5p expression levels in ccRCC. Moreover, these results were mostly corroborated in IPO Porto ccRCC cohort #1, also confirming previous reports [28, 29], and providing compelling evidence that miR-30a-5p downregulation in ccRCC might be caused by aberrant promoter methylation. Thus, our results add miR-30a-5p to the growing list of epigenetically-deregulated microRNAs in urologic malignancies [22,23,24,25], reinforcing the contribution of epigenetic alterations to renal carcinogenesis.
Notwithstanding the mechanism underlying miR-30a-5p downregulation in ccRCC, our study firstly demonstrated that miR-30a-5pme levels might be a specific biomarker for this cancer type. Indeed, since high methylation levels are cancer-specific, they may be used as a tool for ccRCC identification, both in tissue (e.g., as an ancillary tool for histopathological or cytopathological workup of renal mass) and urine samples, providing, in the latter case, a non-invasive tool for early disease detection in high-risk populations [30, 31] (e.g., patients with end-stage chronic renal disease undergoing haemodialysis). Although other hypermethylated miRs (miR-9, miR-124-3) have been proposed as molecular biomarkers for ccRCC [32, 33], their performance in urine samples has not been assessed, yet. Thus, to the best of our knowledge, this is the first miR methylation-based urine biomarker to be proposed for ccRCC. Importantly, it should be emphasized that miRme assessment has several advantages, including higher stability, reduced amount of clinical material requirements and methodological celerity compared to RNA expression assays. Thus, methylation analysis is more robust, enabling the development of tests for use in clinical practice [34, 35].
The rising incidence of incidentally detected RCC presents a significant clinical challenge owing to the risk of overtreatment. Thus, perfecting prognostic models through the inclusion of molecular biomarkers might contribute to reduce that risk. Remarkably, we demonstrated that higher miR-30a-5pme levels assessed in tissue samples independently predicted shorter time to relapse, showing promise as biomarker for risk-stratification among ccRCC and more accurate identification of the high-risk patient subset which may require alternative therapeutic interventions.
Although miR-30a-5pme biomarker performance in Cohort #3 was not impressive, it should be highlighted that miR-30a-5pme levels were able to identify six out of each ten ccRCC in urine samples and, notably, correctly classified seven out of each ten suspects. This simple and cost-effective method is likely to increase patient compliance while reducing the risk of mistreatment. Moreover, since metastatic patients disclosed significantly higher miR-30a-5pme levels than non-metastatic ccRCC, this non-invasive test might also provide relevant information concerning patient monitoring after curative-intent surgery.
The main limitation of our study is the relatively small number of urine samples tested. Moreover, accuracy might be improved by adding additional markers to the panel, as we previously demonstrated for other urologic cancers [22,23,24,25]. Nonetheless, the novelty of using a miRNA methylation marker with diagnostic and prognostic value, amenable for non-invasive detection, constitutes, in our view, a relevant contribution to the field and, hopefully, will stimulate the design of validation studies in larger and independent series.
Conclusions
We demonstrated that miR-30a-5p downregulation, probably due to aberrant promoter methylation, is common in ccRCC. Importantly, miR-30a-5pme levels might aid for diagnostic and prognostic purposes, helping to identify patients at higher risk for disease progression and metastization, although additional markers should be included to improve its overall performance as diagnostic/prognostic biomarker.
Availability of data and materials
The datasets analysed during the current study are available in the TCGA repository (http://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp).
Change history
15 August 2022
A Correction to this paper has been published: https://doi.org/10.1186/s13046-022-02450-x
Abbreviations
- AC:
-
Asymptomatic controls
- AUC:
-
Area under the curve
- ccRCC:
-
Clear cell Renal Cell Carcinoma
- DFS:
-
Disease-free survival
- DSS:
-
Disease-specific survival
- MFS:
-
Metastasis-free survival
- miR:
-
MicroRNA
- miR-30a-5pme :
-
MiR-30a-5p promoter methylation
- OS:
-
Overall survival
- qMSP:
-
Quantitative methylation-specific PCR
- RCC:
-
Renal cell carcinoma
- RT:
-
Reverse transcription
- RFS:
-
Recurrence-free survival
- RNT:
-
Renal normal tissue
- ROC:
-
Receiver operator characteristics
- TCGA:
-
The Cancer Genome Atlas
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Protzel CMM, Hakenber OW. Epidemiology, Aetiology, and pathogenesis of renal cell carcinoma. Eur Urol Suppl. 2012;11(3):52–9.
Aguiari G. MicroRNAs in clear cell renal cell carcinoma: biological functions and applications. J Kidney Cancer VHL. 2015;2(4):140–52.
Silva-Santos RM, Costa-Pinheiro P, Luis A, Antunes L, Lobo F, Oliveira J, et al. MicroRNA profile: a promising ancillary tool for accurate renal cell tumour diagnosis. Br J Cancer. 2013;109(10):2646–53.
Ran L, Liang J, Deng X, Wu J. miRNAs in Prediction of Prognosis in Clear Cell Renal Cell Carcinoma. BioMed Res Int. 2017;2017:4832931.
Xing T, He H. Epigenomics of clear cell renal cell carcinoma: mechanisms and potential use in molecular pathology. Chin J Cancer Res. 2016;28(1):80–91.
Kowalik CG, Palmer DA, Sullivan TB, Teebagy PA, Dugan JM, Libertino JA, et al. Profiling microRNA from nephrectomy and biopsy specimens: predictors of progression and survival in clear cell renal cell carcinoma. BJU Int. 2017;120(3):428–40.
Zhang N, Zhou B, Huang Q, Chen X, Cui S, Huang Z, et al. Multiple metastases of clear-cell renal cell carcinoma to different region of the nasal cavity and paranasal sinus 3 times successively: a case report and literature review. Medicine. 2018;97(14):e0286.
He YH, Chen C, Shi Z. The biological roles and clinical implications of microRNAs in clear cell renal cell carcinoma. J Cell Physiol. 2018;233(6):4458–65.
Heinzelmann J, Henning B, Sanjmyatav J, Posorski N, Steiner T, Wunderlich H, et al. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol. 2011;29(3):367–73.
Davis BN, Hata A. Regulation of MicroRNA biogenesis: a miRiad of mechanisms. Cell Comm Signaling. 2009;7:18.
Mlcochova H, Hezova R, Stanik M, Slaby O. Urine microRNAs as potential noninvasive biomarkers in urologic cancers. Urologic Oncol. 2014;32(1):41.e1–9.
Lorenzen JM, Thum T. Circulating and urinary microRNAs in kidney disease. Clin J Am Soc Nephrol. 2012;7(9):1528–33.
Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74.
Jeronimo C, Henrique R. Epigenetic biomarkers in urological tumors: a systematic review. Cancer Lett. 2014;342(2):264–74.
Heinzelmann J, Unrein A, Wickmann U, Baumgart S, Stapf M, Szendroi A, et al. MicroRNAs with prognostic potential for metastasis in clear cell renal cell carcinoma: a comparison of primary tumors and distant metastases. Ann Surg Oncol. 2014;21(3):1046–54.
Heinzelmann J, Arndt M, Pleyers R, Fehlmann T, Hoelters S, Zeuschner P, et al. 4-miRNA score predicts the individual metastatic risk of renal cell carcinoma patients. Ann Surg Oncol. 2019;26(11):3765–73.
Yang X, Chen Y, Chen L. The versatile role of microRNA-30a in human Cancer. Cell Physiol Biochemistry. 2017;41(4):1616–32.
Zheng B, Zhu H, Gu D, Pan X, Qian L, Xue B, et al. MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem Biophys Res Commun. 2015;459(2):234–9.
Sun L, Chen Y, Su Q, Tang X, Liang Y, Che G, et al. Increased plasma miRNA-30a as a biomarker for non-small cell lung Cancer. Med Sci Monitor. 2016;22:647–55.
Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM, et al. Identification of a 4-microRNA signature for clear cell renal cell carcinoma metastasis and prognosis. PLoS One. 2012;7(5):e35661.
Padrao NA, Monteiro-Reis S, Torres-Ferreira J, Antunes L, Leca L, Montezuma D, et al. MicroRNA promoter methylation: a new tool for accurate detection of urothelial carcinoma. Br J Cancer. 2017;116(5):634–9.
Ramalho-Carvalho J, Goncalves CS, Graca I, Bidarra D, Pereira-Silva E, Salta S, et al. A multiplatform approach identifies miR-152-3p as a common epigenetically regulated onco-suppressor in prostate cancer targeting TMEM97. Clin Epigenetics. 2018;10:40.
Ramalho-Carvalho J, Graca I, Gomez A, Oliveira J, Henrique R, Esteller M, et al. Downregulation of miR-130b~301b cluster is mediated by aberrant promoter methylation and impairs cellular senescence in prostate cancer. J Hematol Oncol. 2017;10(1):43.
Torres-Ferreira J, Ramalho-Carvalho J, Gomez A, Menezes FD, Freitas R, Oliveira J, et al. MiR-193b promoter methylation accurately detects prostate cancer in urine sediments and miR-34b/c or miR-129-2 promoter methylation define subsets of clinically aggressive tumors. Mol Cancer. 2017;16(1):26.
Patard JJ, Leray E, Rioux-Leclercq N, Cindolo L, Ficarra V, Zisman A, et al. Prognostic value of histologic subtypes in renal cell carcinoma: a multicenter experience. J Clin Oncol. 2005;23(12):2763–71.
Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, et al. Renal cell carcinoma. Nat Rev Dis Prim. 2017;3:17009.
Huang QB, Ma X, Zhang X, Liu SW, Ai Q, Shi TP, et al. Down-regulated miR-30a in clear cell renal cell carcinoma correlated with tumor Hematogenous metastasis by targeting angiogenesis-specific DLL4. PLoS One. 2013;8(6):e67294.
Wang C, Cai L, Liu J, Wang G, Li H, Wang X, et al. MicroRNA-30a-5p inhibits the growth of renal cell carcinoma by modulating GRP78 expression. Cell Physiol Biochemistry. 2017;43(6):2405–19.
Barros-Silva D, Marques CJ, Henrique R, Jeronimo C. Profiling DNA Methylation Based on Next-Generation Sequencing Approaches: New Insights and Clinical Applications. Genes. 2018;9:9.
Tang Y, Jiang S, Gu Y, Li W, Mo Z, Huang Y, et al. Promoter DNA methylation analysis reveals a combined diagnosis of CpG-based biomarker for prostate cancer. Oncotarget. 2017;8(35):58199–209.
Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W, Tamboli P, et al. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 2010;29(42):5724–8.
Gebauer K, Peters I, Dubrowinskaja N, Hennenlotter J, Abbas M, Scherer R, et al. Hsa-mir-124-3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br J Cancer. 2013;108(1):131–8.
Martins AT, Monteiro P, Ramalho-Carvalho J, Costa VL, Dinis-Ribeiro M, Leal C, et al. High RASSF1A promoter methylation levels are predictive of poor prognosis in fine-needle aspirate washings of breast cancer lesions. Breast Cancer Res Treat. 2011;129(1):1–9.
Costa-Pinheiro P, Montezuma D, Henrique R, Jeronimo C. Diagnostic and prognostic epigenetic biomarkers in cancer. Epigenomics. 2015;7(6):1003–15.
Acknowledgments
The authors are grateful to the Departments of Urology and of Laboratory Medicine of the Portuguese Oncology Institute of Porto for their collaboration in urine collection, in particularly to Berta Reis, Nursing D.
Funding
This work was funded by Research Center - Portuguese Oncology Institute of Porto (Grant PI 74-CI-IPOP-19-2015). Daniela Barros-Silva was supported scholarship from Research Center - Portuguese Oncology Institute of Porto (CI-IPOP-BI-GEBC2018/UID/DTP/00776/POCI-01-0145-FEDER-006868). Elena Aznar is grateful to Ministerio de Educación, Cultura y Deporte of Spain for her José Castillejo Mobility grant (CAS17/00364). CSG was supported by a FCT Fellowship (SFRH/BD/92786/2013). BMC was funded by IF/00601/2012 and NORTE 01–0145-FEDER-000013 grants.
Author information
Authors and Affiliations
Contributions
Study conceptualization and methodology design: RH and CJ; JO and KJ provided ccRCC samples; CSG and BMC did TCGA analysis. GO-P, DB-S, A-IS, EA and MV-C performed experiments. Formal analysis and the original draft preparation: GO-P, DB-S and A-IS; Review and editing: RH and CJ.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by institutional ethics review board (CES-IPOPFG-EPE 518/10). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from the participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: an error was identified in Supplementary Figure 1 where incorrectly labelled graphs were found.
Supplementary information
Additional file 1 Supplementary Figure S1. Expression of miR-30a-5p according to clinicopathological variables in Cohort #1.
Scatter plots of miR-30a-5p expression levels according to metastasis presentation, recurrence and Führman grade (Mann–Whitney U test).
Additional file 2 Supplementary Figure S2. Prognostic value of stage and nuclear grade in Cohort #1. (A)
Disease-free and (B) disease-specific Kaplan-Meier survival curves based on clinicopathological stage and nuclear grade (Log-rank test).
Additional file 3 Supplementary Figure S3. MiR-30a-5p
me levels and prognostic value of stage and nuclear grade in Cohort #2 (A) Scatter plots of miR-30a-5pme levels according to pathological stage (Mann–Whitney U test); (B) Disease-specific and (C) Metastasis-free Kaplan-Meier survival curves based on clinicopathological stage and nuclear grade (Log-rank test).
Additional file 4 Supplementary Table 1.
Univariable and multivariable analysis of clinicopathological and epigenetic variables in TCGA Cohort (OS and RFS). Supplementary Table 2. List of primers’ sequence for qMSP analysis. Supplementary Table 3. qMSP primers’ design within the methylated gene’s sequence. Supplementary Table 4. Cox univariable and multivariable analysis of clinicopathological and epigenetic variables in Cohort #1 (DFS). Supplementary Table 5. Cox univariable analysis of clinicopathological and epigenetic variables in Cohort #1 (DSS). Supplementary Table 6. Cox univariable and multivariable analysis of clinicopathological and epigenetic variables in Cohort #2 (MFS). Supplementary Table 7. Cox univariable analysis of clinicopathological and epigenetic variables in Cohort #2 (DSS).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Outeiro-Pinho, G., Barros-Silva, D., Aznar, E. et al. MicroRNA-30a-5pme: a novel diagnostic and prognostic biomarker for clear cell renal cell carcinoma in tissue and urine samples. J Exp Clin Cancer Res 39, 98 (2020). https://doi.org/10.1186/s13046-020-01600-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13046-020-01600-3