Abstract
Background
Conventional cytogenetic analysis using G-band karyotyping has been the method of choice for prenatal diagnosis, accurately detecting chromosomal abnormalities larger than 5 Mb. However, the method is inefficient for detecting the submicroscopic deletions and duplications that are associated with malformations and mental retardation. This study evaluated the results of the multiplex ligation-dependent probe amplification (MLPA) P245 assay used for prenatal diagnosis in cases with unusual ultrasonographic findings or specifically where parents wanted to be tested. The objective was to compare the results from MLPA with those from conventional cytogenetic testing in order to determine their concordance and the additional diagnostic yield of MLPA over G-band karyotyping.
Results
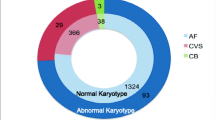
Of the 7522 prenatal cases analyzed, 124 were found to have genomic imbalances (1.6%). Of those 124 cases, 41 had gene loss (33.6%), and 83 had gene gain (66.4%). Most of the cases with genomic imbalances (64.5%) showed no abnormal karyotype. In particular, all cases with a 4p16.3 deletion (Wolf-Hirschhorn syndrome) showed an abnormal karyotype, whereas all of those with a 22q11–13 deletion showed a normal karyotype. In most of the cases with pathogenic deletions, the indication for invasive prenatal testing was an increase in the nuchal translucency (NT) alone (51.2%). Other indications observed in the remaining cases were abnormal serum screening markers (14.6%), other ultrasonographic findings (9.8%), pregnancy through in vitro fertilization and fertility assistance (9.8%), and advanced maternal age(2.4%).
Conclusions
These results show that for fetuses with an enlarged NT or abnormal ultrasonographic findings and normal conventional karyotype, additional genetic investigation like molecular testing would be for identifying the microscopic genomic aberrations (microdeletions, microduplications) responsible for syndromic associations including structural anomalies and mental retardation.
Similar content being viewed by others
Background
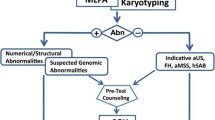
Cytogenetic analysis of fetal cells after chorionic villus sampling or amniocentesis is routinely offered to women who have an increased risk of carrying chromosomally abnormal fetuses. The indications for such prenatal diagnoses include advanced maternal age, increased risk for fetal trisomy identified by maternal serum screening, and fetal abnormalities detected through ultrasonography (USG). In the late first trimester of pregnancy, nuchal translucency (NT) is visible at the back of the neck in all fetuses, where an increased NT is often associated with chromosomal anomalies, especially Down syndrome [1, 2]. If the karyotype is normal but the NT is increased, the fetus has an increased risk of a wide range of congenital malformations, from isolated structural defects to genetic syndromes and neurodevelopmental delays to larger dilemmas of congenital anomalies [3,4,5,6,7,8].
The conventional cytogenetic test using G-band karyotyping has been the method of choice for prenatal diagnosis, accurately detecting chromosomal abnormalities larger than 5 Mb [9, 10]. However, it is inefficient for detecting the sub-microscopic deletions and duplications that are often associated with malformations and mental retardation. Multiplex ligation-dependent probe amplification (MLPA) can examine the subtle abnormalities that cannot be detected by conventional G-band karyotyping [11,12,13,14].
This study presents an overview of the results obtained from use of the MLPA P245 assay for prenatal diagnosis in cases with an unusual USG or in specific cases where parents wanted to be tested. The results from MLPA were compared with those from conventional cytogenetic testing in order to determine their concordance and the additional diagnostic yield of MLPA over G-band karyotyping.
Results
Of the 7522 prenatal cases, 124 were found to have genomic imbalances (1.6%). Of those 124 cases, 41 were found to have gene loss (33.6%), whereas 83 cases had gene gain (66.4%) (Tables 1, 2, 3, and 4). Table 1 summarizes the indications for invasive prenatal diagnosis.
Most of the cases with genomic imbalances (64.5%) did not show an abnormal karyotype (Tables 2 and 3). In particular, all cases with a 4p16.3 deletion (Wolf-Hirschhorn syndrome) showed an abnormal karyotype, whereas those with 22q11–13 deletion showed a normal karyotype (Table 2).
In most of the cases with pathogenic deletions, the indication for invasive prenatal testing was an increased NT alone (51.2%); Other indications observed in the remaining cases were abnormal serum screening markers (14.6%), other findings from USG (9.8%), pregnancy through IVF and fertility assistance (9.8%), and advanced maternal age (2.4%) (Table 1). FISH results were available for 38 of 55 cases with pathogenic imbalances (data not shown).
Discussion
Conventional cytogenetics has been the gold standard in prenatal diagnosis for detecting chromosomal abnormalities. At the light microscope level, the genome-wide numerical and structural anomalies can be examined and the resolution of chromosomal abnormalities of 5–10 Mb can be obtained [9, 10]. Indications for such prenatal diagnoses include advanced maternal age, increased risk for fetal trisomy identified by maternal serum screening, and fetal abnormalities (e.g., increased NT) detected through USG. There is much evidence that measurement of the NT alone or as part of a combined test is an excellent screening method for fetal aneuploidies, and an increased fetal NT with a normal karyotype is associated with an increased risk of adverse pregnancy outcomes [3,4,5,6,7,8].
Euploid fetuses with an increased NT may present with structural anomalies, including cardiac defects, diaphragmatic hernias, exomphalos, body stalk anomalies, and skeletal defects;, where certain genetic syndromes, (e.g., congenital adrenal hyperplasia, fetal akinesia, or Noonan syndrome), have been cited as possible causes. These present a huge dilemma in prenatal counseling. In particular, the 22q11.2 deletion syndrome (also known as DiGeorge or velocardiofacial syndrome) is the most common, with a reported prevalence ranging from 1 in 2000 – to 1 in 6000 live births. The frequency appears to exceed 1 in 1000 in referrals for prenatal diagnosis, even when cases with ultrasonographic evidence for fetal abnormalities are excluded. After Down syndrome, 22q11.2 deletion syndrome is the second most common cause of congenital heart disease and is an important consideration whenever a conotruncal cardiac anomaly is identified, in particular Tetralogy of Fallot. Additional developmental disabilities are often also present [15,16,17].
In this study, we present our experience of using the MLPA P245 assay during invasive prenatal diagnosis in 7522 selected cases with abnormal ultrasonographic findings or parents wanting to be tested. As a result, 124 cases were found to have genomic imbalances with 41 cases having gene loss (33.6%), and 83 having gene gain (66.4%) (Tables 2, and 3). The 22q11.2 deletion syndrome (39%) showed the highest frequency of 2.1 cases per 1000, and all had a normal karyotype (Tables 1, and 2). Conversely, all cases of 4p deletion (19.5%) were accompanied by chromosomal structural abnormality, and the same was true for the cases with a 5p deletion (9.8%) except for only one case. Indications were almost increased NT or ultrasonographic findings (Table 2). Most of the cases were found to have fetal malformations, but almost all of them had normal karyotypes. Thus, whereas deletions are clearly associated with clinical significance or genetic disease, in the cases of duplication, the clinical relevance still remains unclear (Tables 3), [18,19,20,21,22].
Five partial deletions of 22q (one deletion of the three probes) were detected (Tables 1, 2, and 3). MLPA analysis using parental DNA showed that one of the deletions was of maternal origin and two were of paternal origin, and they were expected to be consistent with the parent’s phenotype [23]. The other parents did not wish to tested. All cases were confirmed by the FISH method to be normal. FISH has been the gold-standard method for the diagnosis of microdeletion syndrome, but current FISH probes may not be able to detect all 22q11.2 deletions in the critical region; thus, it is possible that some cases that appeared to be false positive were in fact true positive [15,16,17].
As a consequence, in prenatal genetic counseling, clinicians faced with abnormal ultrasonographic finding, but normal karyotyping, find it difficult to predict the outcomes, and to offer proper genetic counseling and clinical management. Furthermore, the anxiety of prospective parents increases while they wait for the results of more highly informative testing [24,25,26]. In this study we used the MLPA P245 assay for our invasive prenatal chromosome test for a number of reasons. MLPA allows for a more comprehensive determination of gene dosages, with increased convenience and a relatively low cost compared with subtelomeric FISH analysis and other molecular methods. Moreover, MLPA can be used to detect micro-deletions that may not be identified by FISH, and it is more accurate for describing the karyotype owing to its delicate probe spacing. Therefore, the combined use of karyotype analysis and MLPA P245 in prenatal diagnosis facilitates greater accuracy and more informative results than those obtained from use of conventional cytogenetic methods only [27,28,29,30,31,32,33,34].
Conclusions
In conclusion, for fetuses with an enlarged NT or abnormal USG and normal conventional karyotype, additional genetic investigations like molecular testing would be beneficial for identifying the microscopic genomic aberrations (microdeletions, microduplications) responsible for syndromic associations including structural anomalies and mental retardation.
Methods
Patients and samples
All samples from the 7522 pregnant Korean women enrolled in this study were received at the Research Center of Fertility & Genetics of Hamchoon Women’s Clinic between April 2010 and December 2017, for prenatal diagnosis using G-band karyotyping and the MLPA P245 assay. Details of the analyzed tissues are given in Table 4.
Diagnostic techniques
Conventional chromosome analysis was performed on chorionic villi in accordance with the standard procedures used for evaluating numerical and structural chromosome aberrations in direct cytotrophoblastic cells preparations and long-term cultures of mesenchymal tissue (GTG-banding, 550 band level). Chromosome analysis using amniotic fluid and fetal blood was also performed according to standard protocols.
Genomic DNA for MLPA was extracted from T25-flask-cultured fetal cells using a QIAamp DNA kit (Qiagen, Hilden, Germany). The MLPA assay used to screen microdeletion syndromes was performed using the SALSA P245 probemix (MRC-Holland, Amsterdam, Netherlands) according to the manufacturer’s instructions. The amplification products were identified and quantified by capillary electrophoresis using a 3130 XL genetic analyzer (Applied Biosystems, Foster City, CA, USA). The data were analyzed with Gene Marker 1.85 software (SoftGenetics, State College, PA, USA).
Fluorescent in situ hybridization (FISH) was used to assess the structure of the disparity imbalance suspected of being pathogenic in the sample. FISH was performed on metaphase spreads on slides according to the manufacturer’s hybridization protocols.
Abbreviations
- FISH:
-
Fluorescent in situ hybridization
- MLPA:
-
Multiplex ligation-dependent probe amplification
- NT:
-
Nuchal translucency
- USG:
-
Ultrasonography
References
Roozbeh N, Azizi M, Darvish L. Pregnancy outcome of abnormal nuchal translucency: a systematic review. J Clin Diagn Res. 2017;11(3):QC12.
Srebniak MI, De Wit MC, Diderich KE, Govaerts LC, Joosten M, Knapen MF, et al. Enlarged NT (≥3.5 mm) in the first trimester – not all chromosome aberration can be detected by NIPT. Mol Cytogent. 2016;9(1):69.
Simson JM, Sharland GK. Nuchal translucency and congenital heart defects: heart failure or not? Ultrasound Obstet Gynecol. 2000;16(1):30–6.
Bilardo CM, Muller MA, Pajkrt E, Clur SA, Van Zalen MM, Bijlsma EK. Increased nuchal translucency thickness and normal karyotype: time for parental reassurance. Ultrasound Obstet Gynecol. 2007;30(1):11–8.
Bilardo CM, Timmerman E, Pajkrt E, van Maarle M. Increased nuchal translucency in euploid fetuses – what should we be telling the parents? Prenat Diagn. 2010;30(2):93–102.
Dommenico RD, Faraci M, Di Prima FAF, Valenti O, Monte S, Giorgio E, Renda E. Increased nuchal translucency in normal karyotype fetuses. J Prenat Med. 2011;5(2):23–6.
Evangelidou P, Sismani C, Ioannides M, Christodoulou C, Koumbaris G, Kallikas I, et al. Clinical application of whole genome array CGH during prenatal diagnosis: study of 25 selected pregnancies with abnormal ultrasound findings or apparently balanced structural aberrations. Mol Cytogenet. 2010;3:24.
Nikenjade M, Haghighi H. Chromosomally and anatomically normal fetuses with increased first trimester nuchal translucency conceived by ICSI. Iran J Radiol. 2015;12(2):e7157.
Socolov D, Socolov R, Gorduza VE, Butureanu T, Stanculescu R, Carauleanu A, et al. Increased nuchal translucency in fetuses with a normal karyotype – diagnosis and management: an observational study. Medicine (Baltimore). 2017;96(29):e7521.
Kan ASY, Lau ET, Tang WF, Chan SSY, Ding SCK, Chan KYK, et al. Whole-genome array CGH evaluation for replacing prenatal karyotyping in Hong Kong. PLoS One. 2014;9(2):e87988.
Slater HR, Bruno DL, Ren H, Pertile M, Schouten JP, Choo KH. Rapid, high throughput prenatal detection of aneuploidy using a novel quantitative method (MLPA). J Med Genet. 2003;40(12):907–12.
Xu Z, Geng Q, Luo F, Xu F, Li P, Xie J. Multiplex ligation-dependent probe amplification and array comparative genomic hybridization analyses for prenatal diagnosis of cytogenomic abnormalities. Mol Cytogenet. 2014;7(1):84.
Yang WX, Pan H, Li L, Wu HR, Wang ST, Bao XH, et al. Analyses of genotypes and phenotypes of ten Chinese patients with wolf-Hirschhorn syndrome by multiplex ligation-dependent probe amplification and array comparative genomic hybridization. Chin Med J. 2016;129(6):672–8.
Zhang J, Ma D, Wang Y, Coa L, Wu Y, Qiao F, et al. Analysis of chromosome 22q11 copy number variation by multiplex ligation-dependent probe amplification for prenatal diagnosis of congenital heart defect. Mol Cytogenet. 2015;8:100.
Angkustsiri K, Leckliter I, Tartaglia N, Beaton EA, Enriquez J, Simon TJ. An examination of the relationship of anxiety and intelligence to adaptive functioning in children with chromosome 22q11.2 deletion syndrome. J Dev Behav Pediatr. 2012;33(9):713–20.
Gross SJ, Stosic M, Mcdonald DM, Bassett AS, Norvez A, Dhammankar R, et al. Clinical experience with single-nucleotide polymorphism based non-invasive prenatal screening for 22q11.2 deletion syndrome. Ultrasound Obetet Gynecol. 2016;47(2):177–83.
Rapacchia G, Lapucci C, Pittalis MC, Youssef A, Farina A. The first case report in Italy of Di George syndrome detected by noninvasive prenatal testing. Case Rep Obstet Gynecol. 2015;2015:813104.
Barber JC, Bunyan D, Curtis M, Robinson D, Morlot S, Dermitzel A, et al. 8p23.1 duplication syndrome differentiated Frome copy number variation of the defensin cluster at prenatal diagnosis in four new families. Mol Cytogenet. 2010;3:3.
Kiiski K, Roovere T, Zordania R, Von Koskull H, Horelli-Kuitunen H. Prenatal diagnosis of 17p13.1p13.3 duplication. Case Rep Med. 2012;2012:840538.
Liu J, Hu H, Ma N, Jia Z, Zhou Y, Hu JH, et al. A de novo duplication of chromosome 9q34-13-qter in a fetus with tetralogy of Fallot syndrome. Mol Cytogenet. 2016;9:54.
Tropeano M, Ahn JW, Dobson RJB, Breen G, Rucker J, Dixit A, et al. Male-biased autosomal effect of 16p13.11 copy number variation in neurodevelopmental disorders. PLoS One. 2013;8(4):e61365.
Yu Y, Chang L, Zhao H, Li R, Fan Y, Qiao J. Chromosome microduplication in somatic cells decreases the genetic stability of human reprogrammed somatic cells and results in pluripotent stem cells. Sci Rep. 2015;15:10114.
Wilson HL, Wong AC, Shaw SR, Tse WY, Stapleton GA, Phelan MC, et al. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40(8):575–84.
Filges I, Kang A, Klug V, Wenzel F, Heinimann K, Tercanli S, et al. Array comparative genomic hybridization in prenatal diagnosis of first trimester pregnancies at high risk for chromosomal anomalies. Mol Cytogenet. 2012;5(1):38.
Garcia-Herrero S, Campos-Galindo I, Martinez-Conejero JA, Serra V, Olmo L, Lara C, et al. BACs-on-beads technology: a reliable test for rapid detection of aneuploidies and microdeletions in prenatal diagnosis. Biomed Res Int. 2014;2014:590298.
Srebniak M, Boter M, Oudesluijs G, Joosten M, Govaerts L, Van Opstal D, et al. Application of SNP array for rapid prenatal diagnosis: implementation, genetic counselling and diagnostic flow. Eur J Hum Gnnet. 2011;19(12):1230–7.
Buiting K, Dittrich B, Goss S, Lich C, Farber C, Buchholz T, et al. Sporadic imprinting defects in Prader-Willi syndrome and Angelman syndrome: implications for imprint-switch models, genetic counseling, and prenatal diagnosis. Am J Hum Genet. 1998;63(1):170–80.
Butler MG. Benefits and limitation of prenatal screening for Prader-Willi syndrome. Prenat Diagn. 2017;37(1):81–94.
Honjo RS, Dutra RL, Furusawa EA, Zanardo EA, Costa LS, Kulikowski LD, et al. Williams-Beuren syndrome: a clinical study of 55 Brazilian patients and the diagnostic use of MLPA. Biomed Res Int. 2015;2015:903175.
Hu P, Meng L, Ma D, Qiao F, Wang Y, Zhou J, et al. A novel 11p13 microdeletion encompassing PAX6 in a Chinese Han family with aniridia, ptosis and mental retardation. Mol Cytogent. 2015;8(1):3.
Kocks A, Endele S, Heller R, Schroder B, Schafer HJ, Stadtler C, et al. Partial deletion of 4p and 4q in a fetus with ring chromosome 4: phenotype and molecular mapping of the breakpoints. J Med Genet. 2002;39(5):E23.
Sanchez J, Pecina A, Alonso-Luengo O, Gonzalez-Meneses A, Vazquez R, Antinolo G, et al. Atypical association of Angelman syndrome and Klinefelter syndrome in a boy with 47,XXY karyotype and deletion 15q11.2-q13. Case Rep Genet. 2014;2014:517091.
Rince M, Rados M, Krsnik Z, Gotovac K, Borovecki F, Liehr T, et al. Complex intrachromosomal rearrangement in 1q leading to 1q32.2 microdeletion: a potential role of SRGAP2 in the gyrification of cerebral cortex. Mol Cytogenet. 2016;9:19.
Yatsenko SA, Brundage EK, Roney EK, Cheung SW, Chinault C, Lupski JR. Molecular mechanisms for subtelomeric rearrangements associated with the 9q34.3 microdeletion syndrome. Hum Mol Genet. 2009;18(11):1924–36.
Acknowledgements
We would like to thank the family of our patient for their assistance with the clinical evaluation.
Funding
This study was not supported by any foundation.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Detailed clinical information of the patients reported in this paper are however available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
DSL carried out cytogenetic and molecular cytogenetic testing, performed data analysis and draft the manuscript. SHN, SRP and SHG participated in molecular testing. JYM, SHY, KCK and DYH provided samples. SGL and DYH participated in data analysis and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by Hamchoon Women’s Clinic IRB(79507–201,809-BR-002).
Consent for publication
Not applicable.
Competing interests
Authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lee, D., Na, S., Park, S. et al. Clinical experience with multiplex ligation-dependent probe amplification for microdeletion syndromes in prenatal diagnosis: 7522 pregnant Korean women. Mol Cytogenet 12, 10 (2019). https://doi.org/10.1186/s13039-019-0422-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-019-0422-8