Abstract
Background
Cytokinins play many essential roles in plant growth and development, mainly through signal transduction pathways. Although the cytokinin signaling pathway in rice has been clarified, no synthetic reporter for cytokinin signaling output has been reported for rice. The sensitive synthetic reporter two-component signaling sensor (TCSn) is used in the model plant Arabidopsis; however, whether the reporter reflects the cytokinin signaling output pattern in rice remains unclear.
Results
Early-cytokinin-responsive type-A OsRR-binding element (A/G)GAT(C/T) was more clustered in the 15 type-A OsRRs than in the 13 control genes. Quantitative polymerase chain reaction analysis showed that the relative expression of seven type-A OsRRs in roots and shoots was significantly induced by exogenous cytokinin application, and that of seven OsRRs, mainly in roots, was inhibited by exogenous auxin application. We constructed a transgenic rice plant harboring a beta-glucuronidase (GUS) driven by the synthetic promoter TCSn. TCSn::GUS was expressed in the meristem of germinated rice seed and rice seedlings. Furthermore, TCSn::GUS expression in rice seedlings was induced specifically by exogenous cytokinin application and decreased by exogenous auxin application. Moreover, no obvious reduction in GUS levels was observed after three generations of selfing of transgenic plants, indicating that TCSn::GUS is not subject to transgene silencing.
Conclusions
We report here a robust and sensitive synthetic sensor for monitoring the transcriptional output of the cytokinin signaling network in rice.
Similar content being viewed by others
Background
The plant phytohormone cytokinin is a key growth regulator involved in regulating a wide range of developmental processes in diverse contexts throughout the plant life cycle [1,2,3,4]. Cytokinin regulates cell division mainly through histidyl–aspartyl phosphorelay signal transduction (also known as two-component systems), which consist of a histidine protein kinase that senses the input and a response regulator that mediates the output-control signal transduction pathways in many prokaryotes and in some eukaryotes [5,6,7,8]. The Arabidopsis cytokinin signal transduction pathway consists of four principal steps: histidine protein kinase (HK) sensing and signaling; transfer of the phosphoryl group to histidine phosphotransfer protein (HP); response regulator (RR)-dependent transcription activation; and a negative feedback loop through cytokinin-inducible RR genes [4, 6, 9,10,11,12,13,14,15,16].
Although the cytokinin multistep phosphorelay signal transduction pathway is present in Arabidopsis, the identification and functional characterization of the diverse signaling locales is challenging. The distribution of active cytokinin ligands in plants is difficult to determine, mainly due to the synthesis of the cytokinins involved in the complex enzymatic biosynthetic pathways in different cellular compartments [17] and the low level of precise localization (about 100-fold lower than auxin levels) with derived metabolites and different forms [18,19,20]. Cytokinin signaling output at the transcriptional level in Arabidopsis has been visualized using a synthetic reporter, revealing the sites of cytokinin action during wildtype development [21, 22]. Nuclear type-B RRs mediate transcriptional activation in response to phosphorelay signaling activity, whereas type-A RRs repress signaling in a negative-feedback loop. The DNA-binding domains of diverse type-B RR family members are conserved and bind a common DNA-target sequence (A/G)GAT(T/C) in vitro [23,24,25]. Accordingly, the concatemerized type-B ARR-binding motifs are core components of the synthetic reporter [21, 22].
Rice (Oryza sativa L.) is not only a major staple food worldwide but also an important model plant for monocot species research because of its small genome and the availability of its complete genome sequence [26, 27]. The cytokinin signaling pathway in rice has been clarified; however, no synthetic reporter for cytokinin signaling output in rice has been reported. The sensitive synthetic reporter two-component signaling sensor (TCSn) is used in the model plant Arabidopsis; however, whether the reporter reflects the cytokinin signaling output pattern in rice remains unclear. In this study, we analyzed the DNA-binding domains (5′-(A/G)GAT(T/C)-3′) in the promoters of type-A RR family members and examined the changes in the relative expression of type-A RR genes in response to phytohormones. The stronger version of TCSn::GUS was fused into rice plants through genetic transformation. We investigated GUS expression in different generations of transgenic rice seedlings, including in response to exogenous application of six cytokinin fractions and three other phytohormones. We report here a robust and sensitive synthetic sensor for monitoring the transcriptional output of the cytokinin signaling network in rice.
Results and discussion
Amino acid alignment of the Myb-like domains from type-B response regulators
We first selected and analyzed 10 rice type-B positive regulators from the phylogenetic tree described by Tsai et al. [28]. As expected (Additional file 1), the amino acid residues of the C-terminal Myb-like domain [SH(A/L)QKY(R/F)] responsible for their DNA-binding specificity are conserved between rice and Arabidopsis, which is consistent with online registration of a conserved Myb-like motif (accession number: TIGR01557, http://www.jcvi.org/cms/home/). Also, the differences in the sequences of the Myb-like domains may result in regulation of different downstream gene sets in different species.
Analysis of the DNA-binding motif in type-A OsRRs
Cytokinins are classic plant hormones that orchestrate plant growth and physiology and affect gene expression in target cells by activating a multistep phosphorelay network. Type-B RRs, acting as transcriptional activators, mediate the final step in the signaling cascade; simultaneously, type-A RRs are immediate–early target genes of activated type-B RR proteins, which establishes a negative feedback loop to the signaling pathway [4, 15, 16, 19, 22, 29].
To identify the rice type-B RR regulator binding motif (5′-(A/G)GAT(C/T)T-3′) in rice, sharing the conserved pathway with Arabidopsis [30,31,32], the sequence including the promoter and part of the transcribed sequence of rice type-A RR genes (OsRR1-15) was analyzed (Additional file 2) [33]. Additionally, 13 genes that showed stable expression and had no relationship with stress, developmental, pharmacological, or physiological treatments [34,35,36], were randomly selected as negative controls. The total length of the 13 control genes (69.7 kb) was similar to that of the 15 rice type-A RR genes (66.9 kb).
We analyzed the frequency of motifs based on total sequence length and gene sequence. In addition to 66.9 kb from the 15 type-A OsRRs and 69.7 kb from the 13 control genes, a 66.9-kb sequence was randomly selected from the NCBI GenBank database for use as negative control sequences (Additional file 2). The motif number, distance between neighboring motifs, and relative orientation of motifs are important for type-B regulator binding. Based on the distance, motifs were separated into two groups: one with a > 6–30-bp distance and another with a 6–30-bp distance. No difference in the total number of motifs, the motif density, or the number of motifs of distance > 6–30 bp was recorded in the sequences of the 15 type-A genes, 13 control genes, and random sequences (P > 0.05); however, a significant difference was detected in the number of motifs with a 6–30-bp distance irrespective of their direction (P < 0.05). Because motif distance is important for type-B regulator binding [22], our data support the notion that the motif functions in transcription factor binding in natural promoters in rice as well as Arabidopsis. Similarly, a markedly higher mean number of motifs with a 6–30-bp distance was observed in the 15 type-A OsRRs than in the 13 control genes. No apparent bias toward tandem or inverse orientation was recorded in rice (Additional file 3), as in Arabidopsis [22]. To summarize, higher clustered motif were more in rice cytokinin signaling targets than in control genes, which supports the functionality of TCSn in rice.
Changes in the relative expression of type-A OsRRs in shoots and roots in response to cytokinin
Cytokinin-dependent induction of type-A RRs is, in part, dependent on transcriptional regulation by type-B RRs [9, 37]. Similar to Arabidopsis [13], the transcripts of seven rice type-A OsRR genes accumulated rapidly both in roots and shoots after 6-BA treatment for 3 h. Among the type-A OsRR genes in rice, OsRR9 and OsRR10 are difficult to distinguish because of their > 99% nucleotide sequence similarity [38]. Of the other six type-A OsRR genes, the expression of OsRR14/15 was too weak to be detected in rice roots and shoots; and OsRR3,11-13 transcript levels did not respond to 6-BA application both in roots and shoots. To evaluate the responses to other phytohormones of the type-A OsRR genes, we also assessed their expression upon exposure of rice plants to exogenous auxin (IAA), gibberellic acid (GA), and abscisic acid (ABA). Interestingly, transcripts of type-A OsRRs responded to exogenous IAA but not GA and ABA. Application of IAA markedly reduced the transcript levels of most type-A OsRR genes in rice roots, with the exception of OsRR5, OsRR8, and OsRR12-13 (Fig. 1). This is consistent with the notion that auxin antagonizes cytokinin output by direct transcriptional activation of feedback repressors of cytokinin signaling [21, 39]. However, most genes in the shoot, the exception being OsRR4, did not respond to IAA application, suggesting that OsRR family genes in shoots and roots respond differently to exogenous auxin application.
Relative expression of type-A OsRR family genes in response to exogenous phytohormones. Analysis was performed on wild-type plants germinated and grown hydroponically for 14 days, followed by 3 h treatment with 6-benzylaminopurine (6-BA, 100 nM), indole-3-acetic acid (IAA, 1 μM), gibberellic acid (GA, 10 μM), and abscisic acid (ABA, 10 μM). Relative mRNA levels were normalized to those of OsACT. Values are mean ± SD of four biological replicates. *P < 0.05 (ANOVA) between the control and the indicated treatments
Furthermore, OsRR6 and OsRR9/10 levels were higher both in shoots and roots in the control group (Additional file 4). Thus, OsRR6 and OsRR9/10 were selected as marker genes in subsequent experiments.
TCSn::GUS expression patterns are consistent with known cytokinin functions in rice seedlings
To generate a universal cytokinin reporter, a synthetic reporter harboring the concatemerized type-B RR-binding motifs has been tested and optimized in vivo [21]. However, this system has the following limitations: TCS-induced expression is weak in certain developmental contexts in which cytokinin signaling is involved, such as in the shoot and in the vasculature. Furthermore, GFP expression in the root meristem of the seedling decreases as the number of generations increases. Accordingly, an improved version of the reporter, TCS new (TCSn), which, compared with TCS, is more sensitive to phosphorelay signaling in Arabidopsis and maize cellular assays while retaining its specificity, has been developed. TCSn includes variations in the number of binding sites, phasing, and identity of flanking nucleotides [22].
To determine whether TCSn functions in monocotyledon rice and clarify the functions of cytokinins, we constructed a transgenic rice plant harboring beta-glucuronidase (GUS) driven by the synthetic promoter TCSn (Additional file 5; [22]). Cytokinins can stimulate cell division [40, 41]; therefore, we first analyzed the expression pattern of TCSn::GUS in rice seedlings, especially in the meristem. Strong GUS expression was detected in 2-day-old germinated seeds, indicating vigorous cell division (Fig. 2A[a]). Over time, GUS expression was found at the top of the coleoptile and was stronger in the primary root with root hair in 4d-old germinated seeds. As expected, GUS was expressed at the tip of the primary root and in the primordia of the lateral roots of 2-week-old rice seedlings (Fig. 2A[e–g]). The transverse section of the stembase with axillary buds also exhibited stronger GUS activity due to the high cell division capacity. Based on the histochemical localization of TCSn::GUS activity (Fig. 2A), the expression of two Type-A regulator genes (OsRR6 and OsRR9/10) in rice was analyzed in the embryo and endosperm at the tip and bottom of the coleoptile and in the root meristem and maturity zones. OsRR6 and OsRR9/10 expression levels were higher in the embryo than in the endosperm, in the tip than the bottom of the coleoptile and in the meristem than the maturity of the root. These findings are in agreement with the changes in TCSn::GUS expression in tissues, suggesting that GUS activity is consistent with the functions of cytokinins [16, 40, 41] (Fig. 2B).
Histochemical localization of TCSn::GUS activity and gene expression in rice transgenic lines. Analysis was performed on transgenic lines germinated and grown hydroponically for 14 days. A (a) A 2-day-old germinated seed; (b–c) coleoptile (b) and primary root with root hair (c) in a 4-day-old germinated seed; (d–g) transverse section of stembase with axillary bud (d), primary root apical meristem (e), lateral root primordium (f), and primary seedling root with lateral root (g) in a 14-day-old seedling. Bars = 1 mm. B Expression of two Type-A regulator genes (OsRR6 and OsRR9/10) in rice in embryo and endosperm, at the tip and bottom of the coleoptile and in the meristem and maturity zones of the root
TCSn::GUS activity in response to exogenous cytokinin
To assess the sensitivity of TCSn::GUS to cytokinin, we grew transgenic rice seedlings hydroponically for 7 days and then treated them with exogenous cytokinin 6-BA (100 nM) for 12 h. GUS expression was notably induced in roots and shoots by 6-BA application at 3 h (Fig. 3a, b). Furthermore, the supply of 6-BA for 6 h increased GUS activity by 20-fold in roots and by sixfold in shoots compared with the controls. These data suggested that the histochemical staining of TCSn::GUS in roots was more pronounced than in shoots, similar to Arabidopsis [22]. Next, we analyzed GUS activity in response to application of 0–1000 nM 6-BA (Fig. 3c, d). Notably, only application of 1 nM 6-BA significantly enhanced GUS activity by fourfold in rice roots, whereas application of 10 nM 6-BA enhanced GUS activity twofold in rice shoots. Moreover, GUS activity in rice roots and shoots increased with increasing 6-BA concentrations. Finally, we evaluated the effect of different cytokinin fractions on GUS activity. The five cytokinin fractions had an effect similar to that of 6-BA (Fig. 3e, f). These findings suggest that TCSn::GUS expression in roots and shoots was increased by exogenous cytokinin application. Furthermore, we analyzed the expression of OsRR6 and OsRR9/10 (Additional file 6) in the root and found that expression of both matched the TCSn::GUS expression (Fig. 3), confirming that GUS activity is consistent with two marker genes expression for the cytokinin responses. Importantly, no reduction in GUS levels was observed after three generations of selfing of transgenic plants (Additional file 7), indicating that expression of TCSn::GUS is not subject to transgene silencing.
TCSn::GUS activity in response to exogenous cytokinins. Analysis was performed on transgenic lines germinated and grown hydroponically for 7 days followed by application of exogenous cytokinins. a, b Staining (a) and quantification (b) of GUS expression in response to application of 100 nM 6-benzylaminopurine (6-BA) for 12 h; c, d staining (c) and quantification (d) of GUS expression according to 6-BA concentration; e, f staining (e) and quantification (f) of GUS expression in response to treatment with 100 nM cytokinin fractions (Z, zeatin; ZR, zeatin riboside; KT, kinetin; iP, N6-(Δ2-isopentenyl) adenine; iPA, iso-pentenyl adenosine). Values are mean ± SD of four biological replicates. Bars = 2 cm. *P < 0.05 (ANOVA) between the control and the indicated treatments
Specificity of TCSn::GUS in response to exogenous cytokinin
To determine whether the TCSn synthetic promoter responds specifically to cytokinin, we investigated the change in TCSn::GUS expression in response to three other phytohormones. As expected, GUS expression at the stembase and at the tip of primary root was similar to that of other OsRR genes. IAA treatment decreased GUS expression at the stembase and at the tip of the primary root in rice plants (Fig. 4c, h), which is consistent with the antagonism between auxin and cytokinin [42, 43]. Compared with the control, no GUS expression was observed when GA or ABA was applied (Fig. 4). Therefore, cytokinins specifically activate the synthetic promoter TCSn, whereas auxin, gibberellic acid, and abscisic acid do not.
TCSn::GUS activity in response to exogenous phytohormones. Analysis was performed on transgenic lines germinated and grown hydroponically for 7 days followed by 3 h treatment with 6-benzylaminopurine (6-BA, 100 nM), indole-3-acetic acid (IAA, 1 μM), gibberellic acid (GA, 10 μM), and abscisic acid (ABA, 10 μM). A, B GUS expression at the stembase (A) and primary root tip (B); C quantification of GUS expression. Values are mean ± SD of four biological replicates. Bar = 0.5 mm. *P < 0.05 (ANOVA) between the control and the indicated treatments
Conclusion
In this study, we analyzed the DNA-binding domains 5′-(A/G)GAT(T/C)-3′ in type-A OsRR family members and examined the relative expression of type-A OsRR genes in response to treatment with phytohormones. The strong version of TCSn::GUS was fused into rice plants by genetic transformation. We evaluated GUS expression in transgenic rice seedlings, including differences between generations and responses to exogenous application of cytokinin fractions and three phytohormones. A cytokinin-responsive type-A OsRR-binding element was clustered in the upstream sequences of 15 type-A OsRR genes. TCSn::GUS was expressed in the meristem of germinated rice seeds and seedlings. Furthermore, TCSn::GUS expression was induced specifically by exogenous application of cytokinin in rice seedlings. We report here a robust and sensitive synthetic sensor for monitoring the transcriptional output of the cytokinin signaling network in rice.
Methods
Construction of reporter vectors and transformation
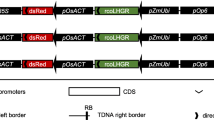
The TCSn promoter sequence [22] was commercially synthesized by the Genewiz Company (Additional file 1). To construct the TCSn::GUS vector, we inserted the synthetic TCSn promoter sequence into the SalI and BamHI sites of the pDR5::GUS vector. The pDR5::GUS construct was kindly provided by the Ping Wu laboratory at Zhejiang University, Hangzhou, China. The construct was transformed into callus initiated from rice seeds (cv. Shiokari) by Agrobacterium tumefaciens (strain EHA105)-mediated transformation. Rice embryonic calli were induced on N6 media, and transformation was performed by Agrobacterium-mediated cocultivation [44].
Plant growth conditions and treatments
Transgenic rice seeds were surface-sterilized with 10% (v/v) H2O2 for 30 min and rinsed thoroughly with deionized water. The sterilized seeds were germinated on a plastic support netting (mesh, 1 mm2) mounted in plastic containers for 1 week. Uniform seedlings were selected and then transferred to a tank containing 7 L of International Rice Research Institute (IRRI) nutrient solution for 2 weeks at pH 5.5. IRRI nutrient solution (1.25 mM NH4NO3, 0.3 mM KH2PO4, 0.35 mM K2SO4, 1 mM CaCl2·2H2O, 1 mM MgSO4·7H2O, 0.5 mM Na2SiO3, 20 μM NaFeEDTA, 20 μM H3BO3, 9 μM MnCl2·4H2O, 0.32 μM CuSO4·5H2O, 0.77 μM ZnSO4·7H2O and 0.39 μM Na2MoO4·2H2O; pH 5.5) was supplied and replaced every 2 days. Plants were grown in a growth chamber at 30 °C during the day and 22 °C during the night with a 16-h light/8 h dark regime. The relative humidity was controlled at ~ 70%.
The cytokinin fractions (6-benzylaminopurine, 6-BA; zeatin, Z; zeatin riboside, ZR; kinetin, KT; N6-(Δ2-isopentenyl) adenine, iP; iso-pentenyl adenosine, iPA), 1 µM indole-3-acetic acid (IAA), 10 µM gibberellic acid (GA), and 10 µM abscisic acid (ABA) were added to the hydroponic media. The seedlings were treated with 100 nM 6-benzylaminopurine (6-BA) at 0, 0.5, 1, 3, and 6 h and with 0, 1, 10, 100, and 1000 nM 6-BA for 6 h.
Quantitative reverse-transcription polymerase chain reaction analysis of gene expression
Total RNA was isolated from the roots of rice seedlings. RNA extraction, reverse transcription, and quantitative reverse-transcription polymerase chain reaction (qRT-PCR) were performed as described previously [45]. Primer sets for type-A OsRRs are listed in Additional file 8.
Histochemical GUS staining and quantitative measurement of GUS activity
Histochemical GUS staining was performed as described previously [46]. The tissues were treated for 1 h with 1.8 M KOH solution heated to 90 °C and then treated with 1% HCl (v/v) solution for 5 min [47]. The stained tissues were photographed using an Olympus SZX2ILLK stereomicroscope equipped with a color charge-coupled device (CCD) camera (Olympus) [48].
For enzymatic GUS assays, tissues were ground in liquid nitrogen and transferred to microtubes containing 1 mL of the extraction buffer (50 mM sodium phosphate, pH 7.5; 10 mM Na2EDTA; 10 mM 2-mercaptoethanol; 0.1% Triton X-100; and 0.1% (w/v) sodium lauryl-sarco-sine) for total protein extraction. Beta-glucuronidase activity was measured fluorimetrically using 1 µg of total protein extract, as described previously [47].
Data analysis
Experimental data were pooled for calculation of means and standard deviations (SDs) and subjected to one-way analysis of variance (ANOVA), followed by least significant difference (LSD) testing to determine the significance of differences between individual treatments. Data in Additional file 3 were pooled and analyzed by nonparametric tests, followed by two independent-samples t tests to determine the significance of differences between two independent samples. All statistical procedures were performed using SPSS version 11.0 (SPSS, Chicago, IL, USA).
References
Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol. 1957;11:118–30.
Mok DW, Mok MC. Cytokinin metabolism and action. Annu Rev Plant Physiol Mol Biol. 2001;52:89–118.
Aloni R, Aloni E, Langhans M, Ullrich CI. Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann Bot. 2006;97:883–93.
Perilli S, Moubayidin L, Sabatini S. The molecular basis of cytokinin function. Curr Opin Plant Biol. 2010;13:21–6.
Hwang I, Chen HC, Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002;129:500–15.
Heyl A, Schmülling T. Cytokinin signal perception and transduction. Curr Opin Plant Biol. 2003;6:480–8.
Ferreira FJ, Kieber JJ. Cytokinin signaling. Curr Opin Plant Biol. 2005;8:518–25.
Müller B, Sheen J. Advances in cytokinin signaling. Science. 2007;318:68–9.
Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–9.
Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T. The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 2001;42:107–13.
Suzuki T, Sakurai K, Ueguchi C, Mizuno T. Two types of putative nuclear factors that physically interact with histidine-containing phosphotransfer (Hpt) domains, signaling mediators in His-to-Asp phosphorelay, in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:37–45.
Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell. 2006;18:3073–87.
Du L-M, Jiao F-C, Chu J, Jin G-L, Chen M, Wu P. The two-component signal system in rice (Oryza sativa L.): a genome-wide study of cytokinin signal perception and transduction. Genomics. 2007;89:697–707.
Punwani JA, Kieber JJ. Localization of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin. Plant Signal Behav. 2010;5:896–8.
Argueso CT, Raines T, Kieber JJ. Cytokinin signaling and transcriptional networks. Curr Opin Plant Biol. 2010;13:533–9.
Hwang I, Sheen J, Müller B. Cytokinin signaling networks. Annu Rev Plant Biol. 2012;63:353–80.
Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot. 2008;59:75–83.
Werner T, Köllmer I, Bartrina I, Holst K, Schmülling T. New insights into the biology of cytokinin degradation. Plant Biol. 2006;8:371–81.
Bishopp A, Lehesranta S, Vatén A, Help H, El-Showk S, Scheres B, et al. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr Biol. 2011;21:927–32.
Antoniadi I, Plačková L, Simonovik B, Doležal K, Turnbull C, Ljung K, et al. Cell-type-specific cytokinin distribution within the Arabidopsis primary root apex. Plant Cell. 2015;27:1955–67.
Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453:1094–7.
Zürcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Müller B. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 2013;161(3):1066–75.
Sakai H, Aoyama T, Oka A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 2000;24:703–11.
Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, et al. Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell. 2002;14:2015–29.
Imamura A, Kiba T, Tajima Y, Yamashino T, Mizuno T. In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:122–31.
Feng Q, Zhang Y-J, Hao P, Wang S-Y, Fu G, Huang Y-C, et al. Sequence and analysis of rice chromosome 4. Nature. 2002;420:316–20.
Sasaki T, Matsumoto T, Yamamoto K. The genome sequence and structure of rice chromosome 1. Nature. 2002;420:312–6.
Tsai YC, Weir NR, Hill K, Zhang W, Kim HJ, Shiu SH, et al. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012;158(4):1666–84.
Werner T, Schmülling T. Cytokinin action in plant development. Curr Opin Plant Biol. 2009;12(5):527–38.
D’Agostino IB, Deruère J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000;124:1706–17.
Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A. ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 2007;48:263–77.
Brenner WG, Schmülling T. Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific responses. BMC Plant Biol. 2012;12:112.
Ritter DI, Dong Z, Guo S, Chuang JH. Transcriptional enhancers in protein-coding exons of vertebrate developmental genes. PLoS ONE. 2012;7:e35202.
Jain M, Nijhawan A, Tyagi AK. Khurana. Validation of housing keep genes as internal control for studying gene expression in rice quantitative real-time PCR. Biochem Biophys Res Commun. 2006;345(2):646–51.
Wang Z-H, Wang Y, Yang J, Hu K-K, An B-G, Deng X-L, et al. Reliable selection and holistic stability evaluation of reference genes for rice under 22 different experimental comditions. Appl Biochem Biotechnol. 2016;179(5):753–75.
Auler PA, Benitez LC, do Amaral MN, Vighi IL, Dos Santos Rodrigues G, da Maia LC, et al. Evalution of stability and validation of reference genes for RT-qPCR expression studies in rice plants under deficit. J Appl Genet. 2017;58(2):163–77.
Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, et al. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science. 2001;294:1519–21.
Hu Z, Yamauchi T, Yang J, Jikumaru Y, Tsuchida-Mayama T, Ichikawa H, et al. Strigolactone and cytokinin act antagonistically in regulating rice mesocotyl elongation in darkness. Plant Cell Physiol. 2014;55:30–41.
Chandler JW, Werr W. Cytokinin-auxin crosstalk in cell type specification. Trends Plant Sci. 2015;20:291–300.
Azizi P, Rafii MY, Maziah M, Abdullah SN, Hanafi MM, Latif MA, et al. Understanding the shoot apical meristem regulation:a study of the phytohormones, auxin andcytokinin, in rice. Mech Dev. 2015;135:1–15.
Perilli S, Moubayidin L, Sabatini S. The molecular basis of cytokinin function. Curr Opin Plant Biol. 2010;13(1):21–6.
Nordstrom A, Tarkowski P, Tarkowska D, Norbaek R, Astot C, Dolezal K, et al. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor pf potential importance for auxin cytokinin regulated development. Proc Natl Acad Sci USA. 2004;101:8039–44.
Shimizu-Sato S, Tanaka M, Mori H. Auxin–cytokinin interactions in the control of shoot branching. Plant Mol Biol. 2009;69:429–35.
Ai P-H, Sun S-B, Zhao J-N, Fan X-R, Xin W-J, Guo Q, et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 2009;57:798–809.
Chen Y-N, Fan X-R, Song W-J, Zhang Y-L, Xu G-H. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol J. 2012;10:139–49.
Konishi M, Yanagisawa S. Identification of a nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. Plant J. 2010;63:269–82.
Chen A-Q, Gu M, Sun S-B, Zhu L-L, Hong S, Xu G-H. Identification of two conserved cis-acting elements, MYCS and P1BS, involved in the regulation of mycorrhiza-activated phosphate transporters in eudicot species. New Phytol. 2011;189:1157–69.
Sun H-W, Tao J-Y, Xie X-N, Yoneyama K, Zhang Y-L, Xu G-H, et al. Strigolactones are involved in phosphate and nitrate deficiency-induced root development and auxin transport in rice. J Exp Bot. 2014;65:6735–46.
Authors’ contributions
JY and HW conceived the study. JY and PG carried out the experiments. JY and YZ drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was funded by the National Nature Science Foundation of China (Grant Nos. 31471936 and 31672225), National key R & D projects (2017YFD0200103), Innovative Research Team Development Plan of the Ministry of Education of China (No. IRT1256), the 111 Project (No. 12009), the Priority Academic Program Development of Jiangsu Higher Education Institutions Project, China Scholarship Council (CSC) and Innovative Plan of Jiangsu Province of China (KYZZ16_0380). The Englsih in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see: http://www.textcheck.com/certificate/vXOTa4.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Yes.
Consent for publication
Yes.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
13007_2017_232_MOESM3_ESM.xlsx
Additional file 3. Summary of 5′-(A/G)GAT(T/C)-3′ motifs in rice cytokinin type-A genes, control genes and random sequence.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Tao, J., Sun, H., Gu, P. et al. A sensitive synthetic reporter for visualizing cytokinin signaling output in rice. Plant Methods 13, 89 (2017). https://doi.org/10.1186/s13007-017-0232-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13007-017-0232-0