Abstract
Nanoparticles exhibit a specific diffusion and sedimentation behavior under cell culture conditions as used in nantoxicological in vitro testing. How a particular particle suspension behaves depends on the particular physicochemical characteristics of the particles and the cell culture system. Only a fraction of the nanoparticles applied to a cell culture will thus reach the cells within a given time frame. Therefore, dosimetric calculations are essential not only to determine the exact fraction of nanoparticles that has come into contact with the cells, but also to ensure experimental comparability and correct interpretation of results, respectively. Yet, the use of published dosimetry models is limited. Not the least because the correct application of these in silico tools usually requires bioinformatics knowledge, which often is perceived a hurdle. Moreover, not all models are freely available and accessible. In order to overcome this obstacle, we have now developed an easy-to-use interface for our recently published 3DSDD dosimetry model, called NanoPASS (NanoParticle Administration Sedimentation Simulator). The interface is freely available to all researchers. It will facilitate the use of in silico dosimetry in nanotoxicology and thus improve interpretation and comparability of in vitro results in the field.
Similar content being viewed by others
Introduction
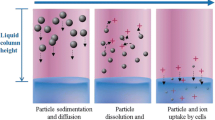
The number of in vitro studies on particle uptake, effects and toxicity is increasing. While the main focus of toxicological research has been in the field of inorganic nanoparticles, plastic particles, especially microplastics, have recently also come into focus [2]. Small-sized particles differ by size, shape, surface coating and agglomeration behavior, and show a particular behavior with respect to their concentration distribution in dispersion: within a given time frame, only a fraction of the applied particles will come into contact with cells located at the bottom of a cell culture vessel [10, 11]. This fraction, termed “effective dose”, “delivered dose” or “target cell dose” depends on the physicochemical characteristics of the particles and of the liquid, e.g. the cell culture medium. Together, these characteristics determine how sedimentation and diffusion affect particle distribution over time [3, 12, 13]. Dosimetry, i.e. the determination of the effective dose of particles that have come into contact with cells in in vitro experiments, is a major issue in the field of particle toxicology and a prerequisite for comparability and correct interpretation of results [7, 13]. A number of models and approaches for dosimetry calculations in silico have been published [4, 6, 8, 9, 14]. Latest result show that that the dose perceived by the cells on the bottom of the well after 24 h of exposure is around 85% lower than the administered nominal media concentration [10]. Usage of these models, however, remains limited. Reasons are the limited public availability of some of the models but also real or perceived hurdles regarding the bioinformatics expertise required for operating the respective software packages and algorithms.
Aim
Therefore, we aimed to develop an easy-to-use user interface (termed NanoPASS – NanoParticle Administration Sedimentation Simulator) for our 3DSDD (3D-sedimentation-diffusion-dosimetry) model that was published recently [1]. The 3DSDD model was designed with respect to the complex growth behavior of differentiated cell models such as Caco-2 or HepaRG. Such cell models need to be cultivated for differentiation for up to several weeks prior to incubation with the particles. The 3DSDD model calculates the effective dose received by the cells, taking into account different cell populations at the bottom and the lateral walls of the culture dish, the latter resulting from continuous growth of cells after having reached confluency at the bottom. Moreover, as basis of the in silico calculations, the 3DSDD model allows using either the hydrodynamic particle diameter as measured by light scattering methods, or the diffusion coefficient as measured by nanoparticle tracking analysis, offering more flexibility than previous dosimetry models [1]. The NanoPASS user interface will enable all users in the field to calculate the effective dose of their specific particles in silico, particularly for subsequent use in vitro. The interface works with the freely available software R and RStudio and can be used without knowledge of programming in R. Moreover, we present a detailed manual how to install and use 3DSDD via NanoPASS. Thereby NanoPASS helps to better implement dosimetry in nanotoxicology and therefore facilitates interpretation and comparison of results obtained with different in vitro models and particles.
How to use the user interface NanoPASS
Both the 3DSDD model and the NanoPASS user interface are freely available for use in research. When using NanoPASS, the user can enter all the parameters into the entry mask of the user interface and is presented with an overview of the results expressed in different graphics.
-
1)
We aimed to give as much help and guidance as possible in order to help readers with their effective nanoparticle dose calculations. When using the NanoPASS interface please proceed as follows:
-
2)
Download the NanoPASS installation guide from the supplementary information (Additional file 1).
-
3)
Follow the instructions to install R (v3.6 or later) and RStudio (v1.2 or later).
-
4)
Download the NanoPASS file from GitHub (https://github.com/falfren/NanoPASS) or Additional file 2.
-
5)
Continue to follow the instructions to install the 3DSDD user interface NanoPASS.
-
6)
Open the interface in your browser. Figure 1a gives an impression of the visual appearance of the user interface. A representative image of the output delivered by NanoPASS is presented in Fig. 1b.
-
7)
The interface is separated into 4 tabs shown at the bottom (“User Input”, “Simulation”, “Visualisation”, “Simulation Data Visualisation” and “Credits”). Use the first tab “User Input” to fill in all necessary information about your particles and the in vitro system (for more details about the necessary information see next section).
-
8)
Go to the next tab “simulation“: Check if your data are listed correctly and chose a memory location.
-
9)
Let the system calculate your particle distribution results choosing “start simulation”.
-
10)
Get an impression of your results in the “Simulation Data Visualisation” tap.
-
11)
Choose the desired simulation time point, particles that hit the ground / the wall should be included and export graphs in .png.
Necessary information for dosimetry calculations
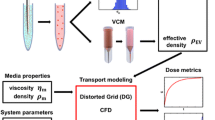
Knowledge of a number of parameters of the nanoparticles, the cell culture dish, and the cell culture medium is necessary to calculate the effective particle dose with the 3DSDD model and the NanoPASS user interface. These parameters can be experimentally determined or estimated. For an overview see Table 1 and the following text or refer to our recent publication [1]. The parameters that are needed refer to different parts of the system that is modeled, namely the characteristics of the nanoparticles, the dimensions of the cell culture dish used for the experiment, characteristics of the cells, characteristics of the cell culture medium and parameters for the calculation model itself.
Nanoparticle characteristics
3DSDD calculates nanoparticles as spherical particles based on size distribution. Prerequisite thus is the determination of the hydrodynamic size in the respective cell culture medium. Please note that measuring in other fluids than the one used for cell incubation may influence the results as the composition of the dispersion medium has an influence on dispersion stability, particle aggregation, ion release and hydrodynamic diameter, respectively. Also, the software will relate to the diameter, not the radius.
When using NTA (Nanoparticle Tracing Analysis) to determine the size of the particles, one can use the diffusion coefficient instead of the hydrodynamic diameter size distribution. As NTA tracks single particles and calculates the hydrodynamic diameter via the diffusion coefficient this actually omits the conversion of the hydrodynamic diameter back into the diffusion coefficient otherwise performed by 3DSDD. An example for particle distribution data is included within the R-package and is further described in the package documentation.
If one does not have the possibility to measure a whole distribution of the particle size or diffusion coefficient, it is also possible to use an average value for the calculation. This will, however, reduce the accuracy of the in silico results.
In addition to the particle size, the software will also require input of the particle density. This is due to the fact that most particles will form agglomerates in cell culture medium and the density allows compensating for any medium inclusions thereof. The effective density therefore needs to be determined in the cell culture medium used for incubation. Merely relying on the material would in most cases cause a massive overestimation of particle sedimentation. Determination of agglomerate density can, for example, be done by analytical centrifugation. Although not common in many toxicological labs, the effective density could also be experimentally estimated by using the PCV tube centrifugation [3]. However, it should be kept in mind that this will provide an average value for the density of the particle agglomerate distribution.
For buoyant particles that have a smaller density then the surrounding fluid and therefore do not sediment to the bottom but float to the surface, the model automatically calculates the floating instead of the sedimentation when the effective density of the particles is smaller than the density of the cell culture medium.
Cell culture dish
To give a correct 3D calculation of the nanoparticle distribution during incubation, the used format of the incubation well and height of medium fluid column needs to be measured. This can be done by using the information provided by the manufacturer, or simply by measuring the size of the cavities of the incubation plate. It has to be kept in mind that the size of cell culture dishes may vary between manufacturers. Medium fluid column height can also be measured or calculated from the well size and medium volume. Table 2 shows some examples.
Cell system
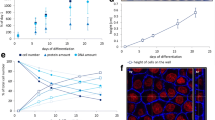
Most adherent cell culture systems are incubated for 24 h post seeding, so that the cells have time to attach to the bottom of the cell culture dish. In these cases the dish bottom area is the area of interest. This parameter was already needed to determine the dimensions of the cell culture dish. Please note there some cell culture models might need longer cultivation times prior to incubation in order to allow for cell differentiation. This is the case, for example, for the most common cell model of the intestinal barrier, Caco-2, as well as for the hepatic cell line HepaRG. After reaching confluency Caco-2 cells need up to 21 days of differentiation to express typical intestinal properties such as the microvilli brush border or tight junctions. The respective monolayer typically reaches a height of about 0.54 cm at the walls of the culture dish. Contrastingly HepaRG reach approximately only half of that (about 0.28 cm) following 4 weeks of differentiation. The cell fraction at the walls is of interest especially for nanoparticle incubation, because these cells come in contact with a different amount of particles compared to the fraction at the bottom. Especially in the commonly used 96-well plate format the number of Caco-2 cells at the wall is much greater than that of cells on the bottom.
Cell culture medium
Likewise some parameters of the incubation medium are needed also for modeling. These include the viscosity and density at the temperature during particle exposure as well as during particle incubation. Especially the density and viscosity will differ slightly with changing medium composition or depending on the additives used. Table 3 provides a few examples.
Calculation parameters
Eventually, three parameters for the calculation need to be defined, namely the number of particles simulated, simulation time, and the sampling interval when snapshots are taken. These parameters can be varied. The more particles are simulated the more detailed are the results, but the more computational resources are needed. A number of 10,000 particles is a good compromise as shown in our previous publication [1]. The simulation time should fit your incubation time on your in vitro experiment. Finally, the sampling interval determines how often within an hour the current position of the simulated particles shall be recorded in the save file. These data shall then be used to calculate the interactive graphical interpretation of the three dimensional in silico particle distribution. A value of 0.25 indicates sampling every 15 min over simulation period defined. However, the smaller the fraction the more snapshots will be taken and the graphical representation will be more fluent. As a downside, the resulting file will increase in size, which may cause memory issues on less powerful machines.
Availability of data and materials
The in silico model 3DSDD can be found in our previous publication [1]. The user interface NanoPASS presented in this short report can be found in the supplementary information.
Abbreviations
- 3DSDD:
-
3D Sedimentation Diffusion Dosimetry
- DLS:
-
Dynamic Light Scattering
- DMEM:
-
Dulbecco’s Modified Eagle’s Medium
- DMSO:
-
Dimethylsulfoxid
- NanoPASS:
-
NanoParticle Administration Sedimentation Simulator
- NTA:
-
Nanoparticle Tracing Analysis
References
Böhmert L, Konig L, Sieg H, Lichtenstein D, Paul N, Braeuning A, Voigt A, Lampen A. In vitro nanoparticle dosimetry for adherent growing cell monolayers covering bottom and lateral walls. Part Fibre Toxicol. 2018;15:42.
Bouwmeester H, Hollman PCH, Peters RJB. Potential health impact of environmentally released micro- and Nanoplastics in the human food production chain: experiences from Nanotoxicology. Environmental Sci Technol. 2015;49:8932–47.
Deloid G, Cohen JM, Darrah T, Derk R, Rojanasakul L, Pyrgiotakis G, Wohlleben W, Demokritou P. Estimating the effective density of engineered nanomaterials for in vitro dosimetry. Nat Commun. 2014a;5:3514.
Deloid GM, Cohen JM, Pyrgiotakis G, Pirela SV, Pal A, Liu J, Srebric J, Demokritou P. Advanced computational modeling for in vitro nanomaterial dosimetry. Particle and Fibre Toxicology. 2015;12:32.
Freshney RI. Culture of animal cells: A manual of basic technique and specialized applications: Wiley; 2015..
Hinderliter PM, Minard KR, Orr G, Chrisler WB, Thrall BD, Pounds JG, Teeguarden JG. ISDD: A computational model of particle sedimentation, diffusion and target cell dosimetry for in vitro toxicity studies. Part Fibre Toxicol. 2010;7:36.
Kowoll T, Fritsch-Decker S, Diabaté S, Nienhaus GU, Gerthsen D, Weiss C. Assessment of in vitro particle dosimetry models at the single cell and particle level by scanning electron microscopy. J Nanobiotechnol. 2018;16:100.
Liu R, Liu HH, Ji Z, Chang CH, Xia T, Nel AE, Cohen Y. Evaluation of toxicity ranking for metal oxide nanoparticles via an in vitro Dosimetry model. ACS Nano. 2015;9:9303–13.
Mukherjee D, Leo BF, Royce SG, Porter AE, Ryan MP, Schwander S, Chung KF, Tetley TD, Zhang J, Georgopoulos PG. Modeling physicochemical interactions affecting in vitro cellular dosimetry of engineered nanomaterials: application to nanosilver. J Nanopart Res. 2014;16:2616.
Poli D, Mattei G, Ucciferri N, Ahluwalia A. An integrated in vitro-in Silico approach for silver nanoparticle Dosimetry in cell cultures. Ann Biomed Eng. 2020;48(4):1271–80.
Rivera GILP, Oberdörster G, Elder A, Puntes V, Parak WJ. Correlating Physico-chemical with toxicological properties of nanoparticles: the present and the future. ACS Nano. 2010;4:5527–31.
Simkó M, Nosske D, Kreyling WG. Metrics, dose, and dose concept: the need for a proper dose concept in the risk assessment of nanoparticles. Int J Environ Res Public Health. 2014;11:4026–48.
Teeguarden JG, Hinderliter PM, Orr G, Thrall BD, Pounds JG. Particokinetics in vitro: Dosimetry considerations for in vitro nanoparticle toxicity assessments. Toxicol Sci. 2006;95:300–12.
Thomas DG, Smith JN, Thrall BD, Baer DR, Jolley H, Munusamy P, Kodali V, Demokritou P, Cohen J, Teeguarden JG. ISD3: a particokinetic model for predicting the combined effects of particle sedimentation, diffusion and dissolution on cellular dosimetry for in vitro systems. Part Fibre Toxicol. 2018;15:6.
Acknowledgements
Not applicable.
Funding
This work was supported by the German Federal Institute for Risk Assessment (projects 1322–637 and 1323–102). Open access funding provided by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
FF transferred 3DSDD from MatLab to R and programmed the NanoPASS. LK wrote the 3DSDD in MatLab. VS, LV, MBP and HS tested NanoPASS, gave feedback and contributed to the manuscript. AB and AV contributed to the manuscript. LB had the idea to program a user interface and contributed to the manuscript. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Frenzel, F., König-Mattern, L., Stock, V. et al. NanoPASS: an easy-to-use user interface for nanoparticle dosimetry with the 3DSDD model. Part Fibre Toxicol 17, 45 (2020). https://doi.org/10.1186/s12989-020-00368-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12989-020-00368-w