Abstract
Background
Encephalomyocarditis virus (EMCV) can cause myocarditis, respiratory failure, reproductive failure, and sudden death in pre-weaned piglets, which has been isolated in China. EMCV VP1 protein was one of the most important structural proteins and played an important role in the protective immunity. In this study, 10 monoclonal antibodies (McAbs) against EMCV VP1 were screened and identified.
Results
Epitope mapping results indicated that McAbs (6E11, 7A7, 7C9) specifically recognized the linear epitopes V(2)ENAEK(7), McAbs (1D1, 2A2, 5A1, 5A11, 5G1) recognized the epitope F(19)VAQPVY(25), and McAbs 1G8 and 3A9 recognized P(42)IGAFTVK(49). Protein sequence alignment of VP1 with 16 EMCV isolates indicated that the epitope F(19)VAQPVY(25) was conserved in all the reference strains. The epitopes P(42)IGAFTVK(49) and V(2)ENAEK(7) only had 1 or 2 variable amino acid among the reference strains. The 3D model analysis results showed that these epitopes presented as spheres were shown within the context of the complete particle.
Conclusions
In this study, ten McAbs against EMCV VP1 were developed and three B-cells epitopes (2-7aa, 19-25aa and 42-49aa) were defined in VP1. All the results herein will promote the future investigations into the function of VP1 of EMCV and development of diagnostic methods of EMCV.
Similar content being viewed by others
Background
Encephalomyocarditis virus (EMCV) is characterized by not only myocarditis and encephalitis, but also neurological diseases, reproductive disorders and diabetes in many mammalian species [1]-[8]. Rodents are considered to be natural hosts of EMCV and are thought to be the primary reservoir and disseminators of the virus. Out of all domestic animals, pigs are the most susceptible to EMCV infection, which can cause severe economic losses on pig production due to high mortality in piglets as a result of respiratory failure [2],[9],[10] and in sows as a result of myocarditis and reproductive failure [11]-[13].
EMCV is a member of the Cardiovirus genus of Picornaviridae[14] and has a single-stranded positive-sense RNA of approximately 7.8 kb [15]. The ORF encodes for a polyprotein that comprises both non-structural and structural elements divided into three primary precursor molecules, namely P1, P2 and P3, encoding for 11 distinct proteins. The structural proteins VP4, VP2, VP3 and VP1 make up the viral capsid and are encoded in the P1 region towards the 5′-end of the genome. The viral capsid proteins, in their capacity to interact with cellular receptors, are crucial for this entry step and may be considered to be factors that can modulate EMCV virulence [1]. The three major capsid proteins, VP1, VP2 and VP3 that constitute the external virion shell of picornaviruses, are considered to play a pivotal role in viral infection and host recognition [16]. Among them, VP1 is one of the most antigenic and can stimulate the organism to produce neutralization antibody [17],[18].
The detailed analysis of epitopes is important for the understanding of immunological events and for the development of epitope-based marker vaccines and diagnostic tools for various diseases [19]-[21]. In this paper, VP1 protein of EMCV NJ08 strain was expressed by the Escherichia coli system and ten monoclonal antibodies (McAbs) against the recombinant EMCV VP1 were screened and identified. Three linear epitopes (2-7aa, 19-25aa and 42-49aa) were identified in VP1 protein of EMCV.
Results
Development of monoclonal antibodies against VP1 protein
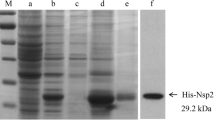
After screening of the fusion cells by indirect ELISA, the positive hybridoma cells secreting the antibodies against VP1 protein were selected and sub-cloned thrice by limiting dilution, and ten McAbs were generated and named as 1D1, 1 F3, 1G8, 1D1, 2A2, 5A1, 5A11, 5G1, 6E11, 7A7 and 7C9. The results of Western blot showed that these McAbs were all directed against the purified EMCV and rVP1 protein expressed in E. Coli (Figure 1). Meanwhile, IFA results also showed that these McAbs could react with the EMCV in BHK21 cells (Figure 1). The titres of antibodies in the cells cultures were 1:1600–3200.
Expression and identification of the truncated fragments
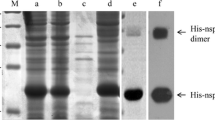
Twenty-six overlapping VP1 protein gene fragments were prepared by PCR and cloned into a GST fusion protein expression vector. After validation by restriction analysis and nucleotide sequencing, recombinant proteins encoded by each of these constructs were expressed in E.coli. The resulting recombinant proteins were identified using Western blot with GST-tagged monoclonal antibody. The results showed that all the proteins were reactive with GST-tagged McAb (Figure 2).
Mapping of the epitopes in VP1
In order to map the minimal sequences of the epitopes recognized by McAbs, the series of truncated recombinant proteins were used in Western blot was to identify the reactivity of each of 10 anti-VP1 McAbs. The results showed that McAbs (6E11, 7A7 and 7C9) could reacted with fragments (F11, F9 and F3) containing the six amino acids V(2)ENAEK(7), but not reacted with the fragments with the deletion of any one of the six amino acids (F10, F7 and F8). It indicated that amino acids V(2)ENAEK(7) in VP1 were essential for this epitope (Figure 3A). In the same way, McAbs (1D1, 2A2, 5A1, 5A11 and 5G1) reacted against the epitope comprising F(19)VAQPVY(25), because they could react with the fragments F15, F13, F18 and F19, but not F12, F16 and F17 (Figure 3B). McAbs (1G8 and 3A9) could reacted with fragments (F25, F26, F21 and F22) containing the eight amino acids P(42)IGAFTVK(49), but not reacted with the fragments with the deletion of any one of the eight amino acids (F24, F23 and F20) (Figure 3C).
Epitope mapping of McAbs by Western blot. The truncated fragments were detected with McAbs. (A) McAb 6B11 specifically reacted with proteins which containing V(2)ENAEK(7) sequence; McAbs 7A7 and 7C9 have the same characteristic as 6E11(data not shown); (B) McAb 1D1 specifically reacted with proteins which containing F(19)VAQPVY(25) sequence; McAbs 2A2, 4B9, 4 F1, 5A1, 5A11 and 5G1 have the same characteristic as 1D1(data not shown); (C) McAb 3A9 specifically reacted with proteins which containing P(42)IGAFTVK(49) sequence; McAb 1G8 has the same characteristic as 3A9(data not shown).
Meanwhile, those purified truncated fragment proteins were used as the antigens in ELISA to detect the levels of those McAbs. The results showed that the OD450 values of McAbs (6E11, 7A7 and 7C9) in F1, F3, F9 and F11 groups were significantly higher than those in F6, F7, F8 and F10 (p < 0.01) (Figure 4A). The OD450 values of McAbs (1D1, 2A2, 5A1, 5A11 and 5G1) in F1, F13, F14, F15, F18 and F19 groups were markedly higher than those in F12, F16 and F17 groups (p < 0.05) (Figure 4B). The levels of McAbs (1G8 and 3A9) in the groups of F21, F22, F25 and F26 were notablely higher than those in F20, F23 and F24 (p < 0.01) (Figure 4C).
Reactivity of McAbs with the different fragments of VP1 expressed in E. coli by ELISA. (A) Reactivity of McAbs, 6E11, 7A7 and 7C9 with the fragments for identifying the epitope 2-7aa. (B) Reactivity of McAbs, 1D1, 2A2, 5A1, 5A11 and 5G1 with the fragments for identifying the epitope 19-25aa. (C) Reactivity of McAbs, 1G8 and 3A9 with the fragments for identifying the epitope 42-49aa.
Confirmation of the epitopes by using peptide-based ELISA
The synthetic peptides (P2-7aa, P19-25 and P42-49aa), purified rVP1 and GST-1-72aa (F3) proteins were used as ELISA antigens. The results showed that OD450 values of 6E11, 7A7 and 7C9 reacted with P2-7aa was highest in those McAbs. And the levels of McAbs (1D1, 2A2, 5A1, 5A11 and 5G1) reacted with P19-25aa were obviously higher than those of other McAbs (P < 0.05). McAbs 1G8 and 3A9 reacted with P42-49aa were significantly higher than other McAbs (P < 0.05). It indicated that the synthetic peptides P2-7aa, P19-25aa and P42-49aa have immune reactivity with the pepide specific McAbs, respectively (Figure 5).
However, the OD450 values of anti-EMCV mice serum were relatively lower than those of McAbs reacted with P2-7aa, P19-25aa and P42-49aa. It suggested that these synthetic peptides reacted weakly with EMCV-specific serum by peptide-based ELISA (Figure 5).
Amino acid differences of the identified epitopes in different EMCV strains
Alignment of the amino acid sequences of the different EMCV isolates revealed that the minimal epitope F(19)VAQPVY(25) was highly conserved among all the reference EMCV strains, the epitope V(2)ENAEK(7) was relatively conserved among all EMCV strains, except for a K7 → R7 change in GXLC, D variant, EMC-B and EMC-D strains. In addition, the epitope P(42)IGAFTVK(49) was conserved among the strains isolated from China. Whereas there were variable amino acids in different site of this region among the reference strains isolated from other countries (Figure 6).
3D model analysis of the three epitopes
There existed a very close structural analog to NJ08 in 2 structures published in 1990 and 1994 with PDB accession numbers 2MEV and 1MEC respectively both for the mengo virus. The VP1 protein of NJ08 had very few amino acid changes and no deletions or additions in the sequence compared to the mengo virus VP1 sequence as shown in Figure 7. Therefore any of the 2 structures could be used a 3D model to map the epitopes within the context of the complete viral particle. As show in Figure 8, the 3 epitopes presented as spheres were shown within the context of the complete particle with colors of red for VP1, green for VP3, yellow for VP2 and blue for VP4. The epitopes V(2)ENAEK(7) and F(19)VAQPVY(25) shown as cyan and magenta spheres were partially buried in the viron. P(42)IGAFTVK(49) shown as orange spheres was located circa the 5-fold icosahedral axis of symmetry and was on the surface.
The locations of the three epitopes. The picture was about the complete particle of mengo virus. VP1 is shown in red ribbon, VP3 is shown in green ribbon, VP2 is shown in yellow ribbon, and VP4 is shown in blue ribbon. The 3 epitopes presented as spheres, V(2)ENAEK(7) is colored cyan, F(19)VAQPVY(25) is colored magenta, and P(42)IGAFTVK(49) is orange. The figure was generated using UCSF chimera [22]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Discussion
B cell epitopes are linear or conformational resulting from unique protein folding. They could be defined as regions on the surface of the native antigen that could bind to B-cell receptors or specific antibodies [21],[23]-[25]. In this study, ten monoclonal antibodies (McAbs) against the VP1 of EMCV were prepared by inoculated with the purified killed EMCV, and then three minimal epitopes [V(2)ENAEK(7), F(19)VAQPVY(25), and P(42)IGAFTVK(49)] were identified in N terminus of the EMCV VP1, using these McAbs and a series of recombinant truncated VP1.
VP1 is one of the most important structures proteins of EMCV, which can stimulate the organism to produce neutralization antibody [18]. Immunization of mice with recombinant VP1 protein (plasmids or recombinant adenoviruses expressing small hairpin RNAs targeted to VP1 protein genes of EMCV), which could provide protective efficacy against EMCV [26],[27]. DNA vaccines encoding an EMCV-D VP1 antigen confer protective immunity against heterologous challenge with EMCV-K, indicating that VP1 is a potential vaccine candidate for controlling EMCV infection [28]. Thus it is need to explore the epitopes related to the protection or diagnosis of this disease, even thought the residue at position 49, 62 and 100 are related to neutralization epitopes. In this study, the ten anti-VP1 McAbs all did not have the ability to neutralize the EMCV in cells cultures (data not shown here). However, they all could react with the EMCV antigen in Western blot and IFA and could be used for diagnosis of this virus.
Sequence analysis of both variants revealed one amino acid exchange within the capsid protein VP1, demonstrated that the diabetogenic and non-diabetogenic EMCV variants differing in only one single amino acid [29]-[32]. Studies of Theiler’s murine encephalomyelitis virus (TMEV) demonstrated that the amino acids on the loops exposed to the surface of the virion were important disease determinants [33]. The residue 49 was located in the loop connecting the βB and βC strands of VP1, which was exposed at the surface of the capsid and was known to be part of a neutralization epitope [34]. In this study, the results of alignment of the amino acid sequences of different EMCV isolates revealed that the epitope F(19)VAQPVY(25) was highly conserved among all the reference EMCV strains. There were only 1 or 2 amino acid residue different in the epitopes P(42)IGAFTVK(49) and V(2)ENAEK(7) between those strains. According to the model of mengo virus, the 3D model of VP1 of EMCV was also analyzed. In the picture, P(42)IGAFTVK(49) shown as orange spheres was located circa the 5-fold icosahedral axis of symmetry and was on the surface. Meanwhile, V(2)ENAEK(7) and F(19)VAQPVY(25) were at the N-terminus and located *under* the VP3 protein and sandwiched with the VP4 protein inside the particle. The obtain of the Mc-Abs against these two epitopes in this study might be explanted by the “breathing” of the N-terminal tail between the VP3 and VP2 proteins. These two epitopes could be used for diagnosis of this virus because of the results of alignment that these two regions were highly conserved among all the reference EMCV strains.
Conclusions
Here, ten McAbs against EMCV VP1 were developed and three B-cells epitopes (2-7aa, 19-25aa and 42-49aa) were defined in VP1. The results may contribute to understand the antigenic structure of VP1 protein deeply and facilitate the development of diagnostic method of EMCV.
Materials and methods
Virus, cells, animals and other reagents
EMCV isolate NJ08 (GenBank Accession No. HM641897) used in this study was isolated and kept in our lab (Submitted 10-JUL-2010). BHK-21 cells were kept in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum. The SP2/0 cells were grown in RPMI 1640 Medium supplemented with 20% fetal bovine serum. Female BALB/c mice, 4–6 weeks of age, were purchased from the Laboratory Animal Centre of Nantong University. HAT and HT Media Supplement (50×) were purchased from Sigma. Horseradish peroxidase (HRP)-tagged goat anti-mouse IgG was purchased from Wuhan Boster Biological Technology Co. Rapid ELISA Mouse McAb Isotyping Kit was purchased from Pierce.
Expression and purification of recombinant VP1 protein in E.coli and the purification of EMCV
The VP1 gene was amplified by PCR using the specific primers kept in our lab. PCR products were digested by Bam H I and EcoR I, and cloned into prokaryotic expression vector pET-28a(+) subsequently. The recombinant plasmids were verified by analysis and nucleotide sequencing, and then transformed into E.coli BL21 cells for fusion proteins expression. The transformants were cultured at 37°C in Luria-Bertani broth containing 0.05 mg/mL kanamycin until the optical density at 600 nm (OD600) reached 0.8. The cells were incubated for another 4 h in the presence of 1.0 mM isopropyl-β-D-thiogalactoside and harvested by centrifugation. The E. coli/pET-28a-VP1 transformants were lysed by sonication, and then centrifuged at 3000 g for 15 min at 4°C. The proteins were purified by ProBondTM Purification System, according to the manufacturer’s instructions (Invitrogen). The purified recombinant VP1 (rVP1) protein was detected by SDS-PAGE and Western blot, and the concentration was determined by NANODROP 2000 Spectrophotometer (Thermo).
EMCV was sedimented and purified from the culture medium with PEG-6000 by ultracentrifugation. The purified EMCV were analyzed by SDS-PAGE and Western blot with anti-EMCV sera.
Development of monoclonal antibodies (McAbs)
The McAbs were developed and obtained as follows. Briefly, inactivated EMCV was combined with equal volumes of complete/incomplete Freund’s adjuvant (Sigma-Aldrich) at final concentration of 0.2 mg/ml. Each female BALB/c mice (6-week-old) was immunized with 0.1 mg emulsion at 3 weeks intervals for 9 weeks. Three days after the last injection, the spleens were surgically removed from the mice and fused with Mouse myeloma cells (SP2/0) using 50% (v/v) PEG. The fused hybridoma clones were screened by the indirect ELISA for McAbs that have strong reactivity with the rVP1 but not His protein or bound EMCV but not BHK-21 cells’ protein. Selected clones were subcloned by limiting dilution. Ascites fluids were induced in pristine-primed BALB/c mice. Finally, the McAbs were identified by Western blot and indirect immunofluorescence assay (IFA).
Indirect ELISA
For screening the McAbs, ELISA plates were coated with the purified rVP1 protein and E.coli/pET-28a lysate (as the negative control) at the concentration of 0.002 mg/mL in buffer bicarbonate (pH9.6) at 4°C overnight or coated with the purified EMCV and BHK-21 cells’ protein at the concentration of 0.01 mg/mL as the same way above. After washing with PBS containing 0.05% Tween-80 (PBST) three times, the plates were blocked with 5% non-fat milk in PBST at 37°C for 2 h. Then wells were incubated with the supernatant of hybridoma at 37°C for 1 h. The sera isolated from immunized mice and the supernatant of SP2/0 cells were used as positive and negative controls, respectively. After three washes with PBST, wells were incubated with a 1:10,000 diluted goat anti-mouse IgG-HRP at 37°C for 1 h. After washing, the plates were incubated with substrate solution tetramethyl benzidine (TMB) at 37°C for 10 min, and the reaction was stopped with 2 M H2SO4 in each well (50 ml/well).The OD was read at 450 nm in an automatic ELISA plate reader. The criteria were judged as follows: the OD450 of the positive control should be more than 1.0, the OD450 of the negative controls should be lower than 0.2. The results were expressed as the ratio of OD450 produced by the samples compared to the negative control. Samples, giving a ratio value higher than 2.1, were considered to be positive.
For mapping the epitopes precisely, a series of truncated fragments of protein were expressed in E.coli. The epitopes recognized by McAbs were mapped by truncated VP1 fragments based on indirect ELISA as described above.
Precise localization of epitopes
In order to identify the minimal antigenic epitopes recognized by McAbs, the VP1 gene was divided into 26 truncated fragments (Figure 9). A series of primers (Table 1) were synthesized to amply the different truncated fragments of VP1 protein. PCR products were separately cloned into the Xho I and Bam H I sites of pGEX-6p-1 expression vector. The recombinant proteins were expressed in E.coli BL21 and identified by SDS-PAGE and Western blot with the GST-tagged monoclonal antibody (Boshide, Wuhan, China), and then were used to determine the epitopes recognized by McAbs using indirect ELISA and Western blot methods.
Western blot
To identify the proteins expressed in E. coli (VP1 and truncated VP1 proteins), the proteins were separated by SDS-PAGE (5-12%) and transferred to nitrocellulose membrane (Pall Corporation). The McAbs (dilution of 1:10–1:50) bound to the proteins on the membrane was detected by goat anti-mouse IgG-HRP (Boshide, Wuhan, China) at 37°C for 1 h. Detection was performed using chemiluminescenceluminol reagents (SuperSignal West Pico Trial kit, PIERCE).
Indirect immunofluorescence assay (IFA)
BHK21 cells infected with EMCV NJ08 strain in 96-well culture plate were rinsed with PBS and fixed with cold ethanol for 45 min at 4°C. The cells were washed, and then incubated with the McAbs (1:10 diluted in PBST) for 1 h at 37°C. After washing with PBST, the cells were incubated with goat anti-mouse IgG conjugated with fluorescein (1:50 diluted in PBST) for 30 min at 37°C. After rinsing by five times, cells were kept in PBS and observed under a ZEISS fluorescence microscope.
Peptide-based ELISA
Three linear epitopes (P2-7aa, P19-25aa and 42-49aa) identified in this study were synthesized using Fmoc solid-phase chemistry (Invitrogen) with >98% purity. They were coated in Reacti-Bind™ Amine-binding, Maleic Anhydride 96-well plates (Pierce, U.S.A.) at the concentration of 10ug/ml for 5 h at room temperature. Meanwhile, purified GST-1-72aa (F3) and rVP1 protein were used as positive antigens control. Mice anti-EMCV serum (at dilution of 1:100) were added as the primary antibody and incubated at 37°C for 1 h. The SPF mice serum was used as negative antibody control. The following detection steps were the same as described above.
Alignment of VP1 epitopes sequences
Nucleotide sequences of EMCV strains from different countries were retrieved from Genbank. The amino acid sequences of identified B-cell epitopes were aligned using DNAstar MegAligen software. The representative EMCV strains were listed in Table 2.
3D model analysis of these epitopes
Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Statistics
The differences in the level of antibodies between different groups were determined by One-way repeated measurement ANOVA and Least significance difference (LSD). Differences were considered statistically significant when p < 0.05.
Abbreviations
- McAbs:
-
Monoclonal antibodies
- ORF:
-
Open reading frame
References
Carocci M, Bakkali-Kassimi L: The encephalomyocarditis virus. Virulence 2012, 3: 351-367. 10.4161/viru.20573
Gelmetti D, Meroni A, Brocchi E, Koenen F, Cammarata G: Pathogenesis of encephalomyocarditis experimental infection in young piglets: a potential animal model to study viral myocarditis. Vet Res 2006, 37: 15-23. 10.1051/vetres:2005041
Grobler DG, Raath JP, Braack LE, Keet DF, Gerdes GH, Barnard BJ, Kriek NP, Jardine J, Swanepoel R: An outbreak of encephalomyocarditis-virus infection in free-ranging African elephants in the Kruger National Park. Onderstepoort J Vet Res 1995, 62: 97-108.
Hubbard GB, Soike KF, Butler TM, Carey KD, Davis H, Butcher WI, Gauntt CJ: An encephalomyocarditis virus epizootic in a baboon colony. Lab Anim Sci 1992, 42: 233-239.
Jones P, Mahamba C, Rest J, Andre C: Fatal inflammatory heart disease in a bonobo (Pan paniscus). J Med Primatol 2005, 34: 45-49. 10.1111/j.1600-0684.2004.00091.x
Juncker-Voss M, Prosl H, Lussy H, Enzenberg U, Auer H, Lassnig H, Muller M, Nowotny N: Screening for antibodies against zoonotic agents among employees of the Zoological Garden of Vienna, Schonbrunn, Austria. Berl Munch Tierarztl Wochenschr 2004, 117: 404-409.
LaRue R, Myers S, Brewer L, Shaw DP, Brown C, Seal BS, Njenga MK: A wild-type porcine encephalomyocarditis virus containing a short poly(C) tract is pathogenic to mice, pigs, and cynomolgus macaques. J Virol 2003, 77: 9136-9146. 10.1128/JVI.77.17.9136-9146.2003
Reddacliff LA, Kirkland PD, Hartley WJ, Reece RL: Encephalomyocarditis virus infections in an Australian zoo. J Zoo Wildl Med 1997, 28: 153-157.
Billinis C, Paschaleri-Papadopoulou E, Psychas V, Vlemmas J, Leontides S, Koumbati M, Kyriakis SC, Papadopoulos O: Persistence of encephalomyocarditis virus (EMCV) infection in piglets. Vet Microbiol 1999, 70: 171-177. 10.1016/S0378-1135(99)00137-6
Denis P, Liebig HD, Nowotny N, Billinis C, Papadopoulos O, O’Hara RS, Knowles NJ, Koenen F: Genetic variability of encephalomyocarditis virus (EMCV) isolates. Vet Microbiol 2006, 113: 1-12. 10.1016/j.vetmic.2005.10.032
Dea S, Bilodeau R, Sauvageau R, Martineau GP: Outbreaks in Quebec pig farms of respiratory and reproductive problems associated with encephalomyocarditis virus. J Vet Diagn Invest 1991, 3: 275-282. 10.1177/104063879100300401
Koenen F, Declercq K, Lefebvre J, Strobbe R: Reproductive failure in sows following experimental-infection with a Belgian Emcv isolate. Vet Microbiol 1994, 39: 111-116. 10.1016/0378-1135(94)90091-4
Zhang GZ, Zhao JX, Wang M: Serological survey on prevalence of antibodies to avian paramyxovirus serotype 2 in China. Avian Dis 2007, 51: 137-139. 10.1637/0005-2086(2007)051[0137:SSOPOA]2.0.CO;2
van Sandwyk JH, Bennett NC, Swanepoel R, Bastos AD: Retrospective genetic characterisation of Encephalomyocarditis viruses from African elephant and swine recovers two distinct lineages in South Africa. Vet Microbiol 2013, 162: 23-31. 10.1016/j.vetmic.2012.08.008
Palmenberg AC, Kirby EM, Janda MR, Drake NL, Duke GM, Potratz KF, Collett MS: The nucleotide and deduced amino acid sequences of the encephalomyocarditis viral polyprotein coding region. Nucleic Acids Res 1984, 70: 2969-2985. 10.1093/nar/12.6.2969
Whitton JL, Cornell CT, Feuer R: Host and virus determinants of picornavirus pathogenesis and tropism. Nat Rev Microbiol 2005, 3: 765-776. 10.1038/nrmicro1284
Kobasa D, Mulvey M, Lee JS, Scraba DG: Characterization of Mengo-Virus Neutralization Epitopes.2. Infection of Mice with an Attenuated Virus. Virology 1995, 214: 118-127. 10.1006/viro.1995.9948
Sin JI, Sung JH, Suh YS, Lee AH, Chung JH, Sung YC: Protective immunity against heterologous challenge with encephalomyocarditis virus by VP1 DNA vaccination: effect of coinjection with a granulocyte-macrophage colony stimulating factor gene. Vaccine 1997, 15: 1827-1833. 10.1016/S0264-410X(97)88856-1
Lin X, Zhao J, Qian J, Mao Y, Pan J, Li L, Peng H, Luo Y, Yan J: Identification of immunodominant B- and T-cell combined epitopes in outer membrane lipoproteins LipL32 and LipL21 of Leptospira interrogans. Clin Vaccine Immunol 2010, 17: 778-783. 10.1128/CVI.00405-09
Peng WP, Hou Q, Xia ZH, Chen D, Li N, Sun Y, Qiu HJ: Identification of a conserved linear B-cell epitope at the N-terminus of the E2 glycoprotein of Classical swine fever virus by phage-displayed random peptide library. Virus Res 2008, 135: 267-272. 10.1016/j.virusres.2008.04.003
Song Y, Zhou Y, Li Y, Wang X, Bai J, Cao J, Jiang P: Identification of B-cell epitopes in the NSP1 protein of porcine reproductive and respiratory syndrome virus. Vet Microbiol 2012, 155: 220-229. 10.1016/j.vetmic.2011.09.018
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE: UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 2004, 25: 1605-1612. 10.1002/jcc.20084
Viudes A, Perea S, Lopez-Ribot JL: Identification of continuous B-cell epitopes on the protein moiety of the 58-kiloDalton cell wall mannoprotein of Candida albicans belonging to a family of immunodominant fungal antigens. Infect Immun 2001, 69: 2909-2919. 10.1128/IAI.69.5.2909-2919.2001
Yin CH, Qin LT, Sun MY, Gao YL, Qi XL, Gao HL, Wang YQ, Jang LL, Wang XM: Identification of a linear B-Cell epitope on avian reovirus protein sigmaC. Virus Res 2013, 178: 530-534. 10.1016/j.virusres.2013.09.028
Yu H, Jiang LF, Fang DY, Yan HJ, Zhou JJ, Zhou JM, Liang Y, Gao Y, Zhao W, Long BG: Selection of SARS-coronavirus-specific B cell epitopes by phage peptide library screening and evaluation of the immunological effect of epitope-based peptides on mice. Virology 2007, 359: 264-274. 10.1016/j.virol.2006.09.016
Bai J, Jiang K, Zeshan B, Wang X, Li Y, Jiang P: Inhibition of encephalomyocarditis virus replication by shRNA targeting 1D and 3AB genes in vitro and in vivo. Virus Genes 2012, 44: 183-190. 10.1007/s11262-011-0684-7
Bai J, Jiang K, Zhang L, Wang X, Li Y, Jiang P: Protective efficacy of adenovirus-mediated small interfering RNAs against encephalomyocarditis virus challenge in mice. J Virol Methods 2012, 185: 204-212. 10.1016/j.jviromet.2012.07.004
Suh YS, Ha SJ, Lee CH, Sin JI, Sung YC: Enhancement of VP1-specific immune responses and protection against EMCV-K challenge by co-delivery of IL-12 DNA with VP1 DNA vaccine. Vaccine 2001, 19: 1891-1898. 10.1016/S0264-410X(00)00443-6
Denis P, Koenen F: Molecular analysis of the capsid coding region of a virulent encephalomyocarditis virus isolate after serial cell passages and assessment of its virulence. Arch Virol 2003, 148: 903-912. 10.1007/s00705-002-0972-6
Jun HS, Kang Y, Notkins AL, Yoon JW: Gain or loss of diabetogenicity resulting from a single point mutation in recombinant encephalomyocarditis virus. J Virol 1997, 71: 9782-9785.
Nelsen-Salz B, Zimmermann A, Wickert S, Arnold G, Botta A, Eggers HJ, Kruppenbacher JP: Analysis of sequence and pathogenic properties of two variants of encephalomyocarditis virus differing in a single amino acid in VP1. Virus Res 1996, 41: 109-122. 10.1016/0168-1702(95)01257-5
Zhu S, Ge XN, Gong XW, Guo X, Chen YH, Yang HC: Alteration of encephalomyocarditis virus pathogenicity due to a mutation at position 100 of VP1. Sci China Life Sci 2011, 54: 535-543. 10.1007/s11427-011-4172-z
Jnaoui K, Minet M, Michiels T: Mutations that affect the tropism of DA and GDVII strains of Theiler’s virus in vitro influence sialic acid binding and pathogenicity. J Virol 2002, 76: 8138-8147. 10.1128/JVI.76.16.8138-8147.2002
Guy M, Chilmonczyk S, Cruciere C, Eloit M, Bakkali-Kassimi L: Efficient infection of buffalo rat liver-resistant cells by encephalomyocarditis virus requires binding to cell surface sialic acids. J Gen Virol 2009, 90: 187-196. 10.1099/vir.0.004655-0
Acknowledgements
This work was supported by grants from the Jiangsu natural science foundation to young researcher (BK20130691), the China agricultural research system foundation (CARS-36), the special fund for agroscientific research in the public interest (201003060–4), the National Natural Science Foundation (31230071), and the Priority Academic Program Development of Jiangsu higher education institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: JB, PJ. Performed the experiments: JB, XHC. Analyzed the data: JB, XHC, KFJ, BZ, PJ. Contributed reagents/materials/analysis tools: JB, XHC, KFJ. Wrote the paper: JB and XHC. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Bai, J., Chen, X., Jiang, K. et al. Identification of VP1 peptides diagnostic of encephalomyocarditis virus from swine. Virol J 11, 226 (2014). https://doi.org/10.1186/s12985-014-0226-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-014-0226-8