Abstract
Objectives
Antigen-induced activation and proliferation of HIV-1-infected cells is hypothesized to be a mechanism of HIV persistence during antiretroviral therapy. The objective of this study was to determine if proliferation of H1N1-specific HIV-infected cells could be detected following H1N1 vaccination.
Methods
This study utilized cryopreserved PBMC from a previously conducted trial of H1N1 vaccination in HIV-infected pregnant women. HIV-1 DNA concentrations and 437 HIV-1 C2V5 env DNA sequences were analyzed from ten pregnant women on effective antiretroviral therapy, before and 21 days after H1N1 influenza vaccination.
Results
HIV-1 DNA concentration did not change after vaccination (median pre- vs. post-vaccination: 95.77 vs. 41.28 copies/million PBMC, p = .37). Analyses of sequences did not detect evidence of HIV replication or proliferation of infected cells.
Conclusions
Antigenic stimulation during effective ART did not have a detectable effect on the genetic makeup of the HIV-1 DNA reservoir. Longitudinal comparison of the amount and integration sites of HIV-1 in antigen-specific cells to chronic infections (such as herpesviruses) may be needed to definitively evaluate whether antigenic stimulation induces proliferation of HIV-1 infected cells.
Similar content being viewed by others
Background
A cure for human immunodeficiency virus type-1 (HIV-1) infection may hinge on understanding the mechanisms that allow HIV-1 to persist during otherwise effective antiretroviral treatment (ART). Studies of HIV-1 C2-V5 env and pol DNA and RNA sequences and integration sites during effective ART have shown multiple identical env and pol templates (which we describe as “monotypic”) [1, 2], an increase in the proportion of monotypic env and pol proviruses over time [2, 3], and a linkage between monotypic env through the 3′ LTR proviral sequences with identical chromosomal integration sites [4], all indicating that HIV-infected cells proliferate. This study addressed our hypotheses that proliferation of HIV-1 infected CD4+ T-cells occurs at least in part by exposure to recall antigens, and that antigen-driven proliferation contributes to sustaining the HIV-1 reservoir. We took advantage of a H1N1 influenza vaccination study in HIV-1-infected pregnant women to explore this hypothesis.
Immunological responses to infections [5] and vaccines [6–8] have been associated with increases in HIV-1 plasma viral load, although the effect of influenza vaccination on plasma HIV-1 RNA has been variable [9–12]. The effect of vaccination on the proviral population has not been explored in detail. Studying responses to H1N1 influenza vaccination in pregnant HIV-infected, ART-treated, women who were vaccinated against H1N1 in last few months of 2009 provided a unique opportunity to explore the effect of recall antigen response on the HIV DNA population. The H1N1 pandemic strain emerged in the first few months 2009, and showed antigenic similarities to a strain that circulated in the human population in the mid 1970s [13]. As this strain presented neo-antigens to those born after the mid-1970s, including the study population of HIV-infected women, the study design presumed that participants with a pre-vaccination pandemic H1N1-specific antibody response had been infected and developed an immune response during the 2009 pandemic. However, because the pandemic H1N1 had approximately 70% homology at the T cell epitope level with recent seasonal H1N1 serotypes, very low pre-vaccination responses were considered due to cross-reactivity. Participants who were not on ART in the first half of 2009 during the H1N1 pandemic and had serologic evidence of infection with H1N1 antigens, were expected to have a recall response to the vaccination. Women who subsequently started ART should have proliferation of influenza-specific CD4+ memory T cells after vaccination, some of which would be latently infected with HIV-1. Increases in proviruses due to T-cell proliferation would cause an increase in the proportion of monotypic sequences in the viral population. In contrast, women on effective ART when they were infected with pandemic H1N1 or without pre-existing H1N1 antibody titers would not be expected to have evidence of proliferation of HIV-infected cells after H1N1 vaccination.
To explore whether a response to a recall antigen leads to proliferation of HIV-1 infected cells, the HIV DNA from PBMC was quantified, multiple HIV-1 DNA genetic sequences were generated from each participant from before and after vaccination, and these sequences were analyzed for evidence of proliferation of HIV-infected cells. Because the timing of HIV diagnosis, ART, primary H1N1 antigen exposure, and subsequent H1N1 vaccination are well defined for this cohort, we had the potential opportunity to investigate the specific effect of antigen-driven cell proliferation on HIV persistence during ART.
Methods
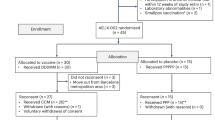
This study utilized demographic and antibody data and specimens from HIV-1 infected pregnant women who had participated in a Phase II Study to Assess the Safety and Immunogenicity of an Inactivated Swine-Origin H1N1 Influenza Vaccine in HIV-1 Infected Pregnant Women (IMPAACT P1086) [14]. As part of P1086, participants’ blood was collected before and 21 days after the first H1N1 vaccination (high dose, 30 μg), at which time (21 days post primary vaccination) they received a H1N1 booster. Participants were selected for this substudy based on: (1) ART-suppression of HIV-1 replication (plasma HIV-1 RNA <50 copies/mL, or plasma HIV-1 RNA <500 copies/mL and decreasing in those who started ART approximately at the same time as enrolling into the study), and (2) specimens available for analysis from before and after vaccination. P1086 study participants were not selected based on pre-existing antibody response to H1N1. H1N1 had entered the US beginning ~9 months prior to the P1086 study, therefore P1086 study enrolled a mixture of participants with and without immunity to H1N1 prior to H1N1 vaccination. Utilizing specimens from this H1N1 vaccination trial allowed a unique opportunity to investigate the HIV population immediately before and after a defined antigen stimulus in a mixed population for which H1N1 was a recall antigen for some individuals and a de novo antigen for others. Study endpoints included (1) PBMC DNA load by limited dilution PCR, (2) proportion of monotypic sequences defined as ≥2 identical HIV C2-V5 env sequences, and (3) quantification of low-level plasma HIV RNA defined as ≥9 c/mL.
DNA was extracted from peripheral blood mononuclear cell (PBMC) samples using a kit (5 Prime ArchivePure DNA Kit; 5 PRIME Inc., Gaithersburg, MD) and quantified using spectrometry (NanoDrop 1000 Spectrophotometer; Thermo Scientific, Waltham, Ma). To quantify HIV-1 DNA, the DNA was serially diluted, triplicates of each dilution underwent PCR to amplify HIV-1 env C2-V5, and the Quality program [15] was used to estimate the number of copies of HIV-1 in the sample, using 1st-round (BH2 [16] and Env6834 (CAG GCC TGT CCA AAA GTA TCC TTT GAG CCA ATT CC), and 2nd-round (DR7 [17] and DR8 [18]) PCR primers, and cycling conditions previously published [19]. To amplify ~20 single viral templates, the DNA from each specimen was diluted to a concentration expected to yield an amplicon from ~30% of PCR; at this concentration 70% of the amplicons are predicted to be from a single viral template. Sequences that appeared in chromatograms to have been derived from more that one template were discarded.
The amplicons were purified (ExoSAP-IT; Affymetrix, Santa Clara, CA) and 2uL of the cleaned product was added to 2uL of primers DR7 and DR8 at concentration of 3.2 pmol for dideoxynucleotide sequencing. Sequences were analyzed using Sequencher program v5.0.1 (Gene Codes Corporation, Ann Arbor, MI) and aligned in Seaview (http://pbil.univlyon1.fr/software/seaview3.html). DIVEIN was used to construct phylogenetic trees and compute divergence of sequences from the most recent common ancestor (MRCA) and sequence diversity [20]. The trees were rooted against reference subtype B sequences except participant 2 who was infected with a complex recombinant and was rooted against a collection of similar sequences in the HIV Sequence Compendium. FigTree (http://tree.bio.ed.ac.uk/software/figtree) was used to graphically represent the phylogenetic trees.
To determine if H1N1 vaccine was associated with low-level viremias, HIV-1 RNA was quantified from subjects’ plasma using previously published methods [21] with the following adaptations. Viral particles from 4.7 mL of plasma (in Beckman OptiSeal Tubes) were pelleted by ultracentrifugation at 175,000 g for 30 min. Afterwards, 3.7 mL of plasma was carefully removed from above the pellet, leaving the 1 mL of plasma overlying the pelleted virions undisturbed. The pellet was resuspended and the HIV RNA was quantified (Abbott RealTime HIV-1 Assay; Abbott Molecular, Abbott Park, Illinois). The calculated lower limit of quantification after concentration of the virions was approximately 9 copies/mL of plasma.
Comparisons of population medians and proportions were performed using Students t-Test for two samples with 2-sided tails (Excel, Microsoft, Redmond, WA).
Results
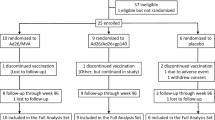
Specimens from 26 participants were processed, but only 10 participants yielded sufficient HIV-1 DNA from before and after vaccination to derive multiple sequences of env. Pre- and post-vaccine data were compared across all 10 participants, and between participants separated into two groups based on whether or not each woman was predicted to exhibit H1N1-specific proliferation of HIV-infected CD4+ T-cells. Group 1 included participants predicted to not have proliferation of HIV-infected cells after vaccination; including those with a negative H1N1 titer pre-vaccination, those without evidence of response to vaccination as measured by Hemagglutination Inhibition assay (HAI), and those on suppressive ART since prior to April 2009, the beginning of the H1N1 pandemic. Group 2 included participants that were predicted to have proliferation of HIV-infected cells based on a pre-vaccine H1N1 Influenza HAI titer ≥1:20, a >three fold increase in post vaccine HAI titer, and initiation of ART in or after April 2009. The Group 2 women would have developed a primary response to the H1N1 pandemic strain before ART initiation. These women were HIV-infected at the time of their H1N1 Influenza infection, and would likely have integrated HIV proviruses in H1N1-specific CD4+ T cells. Based on these criteria, three participants were classified as having potential to show antigen-driven proliferation of HIV-infected H1N1-specific cells in response to vaccination, and seven were not expected to demonstrate proliferation of HIV-infected cells following vaccination. Clinical parameters, including CD4+ cell count, plasma HIV-1 RNA at enrollment, and time on ART are summarized in Table 1.
The study immunization with H1N1 vaccine did not induce significant changes in HIV-1 DNA or RNA concentrations in Group 1 or Group 2 (Table 1). Across the ten women HIV-1 DNA concentrations were similar before and after vaccination (median pre- vs. post-vaccination: 96 vs. 41 copies/million PBMC, p = .37). There was also no change in the percentage of women with detectable low-level HIV RNA (50 vs. 33%, p = .55), among the six women that had ≥4.7 mL of plasma available from before and after vaccination. The change in HIV-1 DNA concentration was not significantly different between the women predicted to have proliferation of HIV-infected cells (median difference post vaccine +4.79 copies HIV-1/million PBMC) versus those that were not (median difference post vaccine −5.73 copies HIV-1/million PBMC; p = .78).
A median of 20.5 (range 18–36) HIV-1 env (C2-V5) sequences were generated from each participant’s pre- and post-vaccine PBMC specimen, with a total of 437 sequences across the ten subjects. Post-vaccine, participants predicted to have H1N1 vaccine-induced proliferation of HIV-infected cells had a median of a 21% decrease in the percentage of monotypic proviral sequences versus a median of a 32% increase in the percentage of monotypic proviral sequences in those not expected to have proliferation of HIV-infected cells after H1N1 vaccination, although this difference was not significantly different (p = .21) (Table 1).
A comparison of HIV-1 DNA sequences from pre- to post-vaccination did not detect differences between the groups. The median change in env sequence divergence from the most recent common ancestor (MRCA) was 0 in Group 1 and .2% in Group 2 (p = .27). The median change in diversity of the env sequences was 0 in Group 1 and .5% in Group 2 (p = .48) (Table 1; Additional file 1).
Discussion
This study looked for evidence that an immunologic recall response by HIV-1 infected T-cells would amplify the HIV-1 proviral reservoir. None of the parameters evaluated demonstrated evidence of antigen-specific proliferation HIV-infected cells following vaccination. Specifically, the participants who were HIV-infected but not on ART during the H1N1 influenza pandemic and had H1N1 immunity prior to H1N1 vaccination did not have an increase in HIV-1 DNA load, proportion of monotypic sequences, or HIV-1 DNA divergence, after H1N1 vaccination.
This was a small study that sought to use existing specimens from a unique cohort that received a timed exposure to a recall antigen. Despite the relatively small size of the study, and the fact that all participants were pregnant women, the results are consistent with another recent study, mostly in men, which did not show a change in HIV DNA load after vaccination [12]. The lack of detectable antigen-driven proliferation in our study suggests that the timing, quantity, or route of antigen exposure by the vaccine was insufficient to alter the HIV proviral population enough to be detected by our methods [12]. We evaluated specimens 21 days after vaccination because that was the time-point at which specimens were collected in the parent study, although peak CD4 T-cell responses may be closer to 10–14 days after vaccination. Despite generating 437 bi-directional HIV env sequences from specimens diluted to single viral templates (a median of 20 individual HIV env sequences per participant before and after vaccination), the power needed to detect antigen-driven proliferation of HIV-infected cells is unknown. A recent study showed increases in HIV transcription after vaccination [12], which suggests that more intensive sampling, perhaps earlier after the exposure to antigen, and/or increased assay sensitivity, may be needed to detect antigen-driven proliferation of HIV-infected cells. Another possibility is that antigen-driven proliferation of HIV-1-infected cells may not have been detected if HAI H1N1 titers were not sensitive or specific for previous H1N1 exposure. Because there were a limited number of cells remaining available from these timepoints and H1N1 CD4 epitopes overlapped with those of other recently circulating influenza strains, we used HAI antibody titers as a proxy for CD4+ T-cell responses. It is possible that CD4 responses to previous influenza infections may not correlate closely enough with measured antibody responses to hemagglutinin. Alternatively, our underlying hypothesis may by wrong, and identical HIV-1 proviruses [2, 4] may arise due to different mechanisms. For example, the proportion of monotypic sequences may increase over time on ART from homeostatic proliferation or dysregulated proliferation of cells with HIV integrated into genes that regulate cell growth.
In summary, a recall antigen, H1N1 influenza vaccine, did not stimulate detectable proliferation of HIV-1 infected cells. This supports the safety of routine influenza vaccination in HIV-1-infected patients on ART. However, to definitively determine whether antigen-driven proliferation of infected cells is a mechanism of HIV persistence, more sensitive methods or alternative study designs may be needed.
Abbreviations
- HIV-1:
-
human immunodeficiency virus type-1
- ART:
-
antiretroviral treatment
- PBMC:
-
peripheral blood mononuclear cell
- H1N1:
-
influenza A virus subtype H1N1
- MRCA:
-
most recent common ancestor
- HAI:
-
hemagglutination inhibition assay
- IMPAACT:
-
international maternal pediatric adolescent aids clinical trials network
References
Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, Haggerty CM, Kamireddi AR, Liu Y, Lee J, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4 + T cells. J Virol. 2006;80:6441–57.
Wagner TA, McKernan JL, Tobin NH, Tapia KA, Mullins JI, Frenkel LM. An increasing proportion of monotypic HIV-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J Virol. 2013;87:1770–8.
Kearney MF, Spindler J, Shao W, Yu S, Anderson EM, O’Shea A, Rehm C, Poethke C, Kovacs N, Mellors JW, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog. 2014;10:e1004010.
Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–3.
Donovan RM, Bush CE, Markowitz NP, Baxa DM, Saravolatz LD. Changes in virus load markers during AIDS-associated opportunistic diseases in human immunodeficiency virus-infected persons. J Infect Dis. 1996;174:401–3.
Stanley SK, Ostrowski MA, Justement JS, Gantt K, Hedayati S, Mannix M, Roche K, Schwartzentruber DJ, Fox CH, Fauci AS. Effect of immunization with a common recall antigen on viral expression in patients infected with human immunodeficiency virus type 1. N E J Med. 1996;334:1222–30.
Brichacek B, Swindells S, Janoff EN, Pirruccello S, Stevenson M. Increased plasma human immunodeficiency virus type 1 burden following antigenic challenge with pneumococcal vaccine. J Infect Dis. 1996;174:1191–9.
Ortigao-de-Sampaio MB, Shattock RJ, Hayes P, Griffin GE, Linhares-de-Carvalho MI, Ponce de Leon A, Lewis DJ, Castello-Branco LR. Increase in plasma viral load after oral cholera immunization of HIV-infected subjects. AIDS. 1998;12:F145–50.
Ho DD. HIV-1 viraemia and influenza. Lancet. 1992;339:1549–0.
Gunthard HF, Wong JK, Spina CA, Ignacio C, Kwok S, Christopherson C, Hwang J, Haubrich R, Havlir D, Richman DD. Effect of influenza vaccination on viral replication and immune response in persons infected with human immunodeficiency virus receiving potent antiretroviral therapy. J Infect Dis. 2000;181:522–31.
Skiest DJ, Machala T. Comparison of the effects of acute influenza infection and Influenza vaccination on HIV viral load and CD4 cell counts. J Clin Virol. 2003;26:307–15.
Yek C, Gianella S, Plana M, Castro P, Scheffler K, Garcia F, Massanella M, Smith DM. Standard vaccines increase HIV-1 transcription during antiretroviral therapy. AIDS. 2016;30:2289–98.
Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N E J Med. 2009;361:1945–52.
Abzug MJ, Nachman SA, Muresan P, Handelsman E, Watts DH, Fenton T, Heckman B, Petzold E, Weinberg A, Levin MJ. Safety and immunogenicity of 2009 pH1N1 vaccination in HIV-infected pregnant women. Clin Infect Dis. 2013;56:1488–97.
Rodrigo AG, Goracke PC, Rowhanian K, Mullins JI. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res Hum Retrovir. 1997;13:737–42.
Altfeld M, Addo MM, Shankarappa R, Lee PK, Allen TM, Yu XG, Rathod A, Harlow J, O’Sullivan K, Johnston MN, et al. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J Virol. 2003;77:7330–40.
Liu SL, Schacker T, Musey L, Shriner D, McElrath MJ, Corey L, Mullins JI. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J Virol. 1997;71:4284–95.
Delwart EL, Shpaer EG, Louwagie J, McCutchan FE, Grez M, Rubsamen-Waigmann H, Mullins JI. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–61.
Tobin NH, Learn GH, Holte SE, Wang Y, Melvin AJ, McKernan JL, Pawluk DM, Mohan KM, Lewis PF, Mullins JI, Frenkel LM. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J Virol. 2005;79:9625–34.
Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, Kosakovsky Pond SL, Mullins JI. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques. 2010;48:405–8.
Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, Dewar RL, Planta A, Liu S, Metcalf JA, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2003;41:4531–6.
Authors’ contributions
TAW and LMF conceived of the project, analyzed the data, and wrote the manuscript. HCH planned and conducted the lab work. CES conducted lab work, assisted in the data analysis, and drafting some sections of the manuscript. KMR identify available specimens and conducted immunological assays. AW and SN assisted in the design of the project and participated in drafting the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We appreciate the support from IMPAACT and thank Petronella Muresan for identifying appropriate specimens.
Partial data presented at Pediatric Academic Societies, San Diego, Apr 25–28, 2015
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All sequences submitted to GenBank, accession numbers X-XX (pending).
Ethics approval
This study conducted using de-identified specimens previously collected as part of IMPAACT P1086, and was approved by The Seattle Children’s IRB (IRB study #13885).
Funding
This work was supported by NIH funds through an International Maternal Pediatric Adolescent AIDS Clinical Trials Group (IMPAACT) Virology Developmental Laboratory award (LMF) (U01 AI068632), a NIH Mentored Patient-Oriented Research Career Development Award to TAW (K23AI077357), and the Clinical Research and Retrovirology Core of the Seattle Centers for AIDS Research (P30 AI027757), and the IMPAACT Statistical and Data Management Center (U01 AI068616). Overall support for IMPAACT was provided by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under Award Numbers UM1AI068632 (IMPAACT LOC), UM1AI068616 (IMPAACT SDMC) and UM1AI106716 (IMPAACT LC), with co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and the National Institute of Mental Health (NIMH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Corresponding author
Additional file
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wagner, T.A., Huang, H.C., Salyer, C.E. et al. H1N1 influenza vaccination in HIV-infected women on effective antiretroviral treatment did not induce measurable antigen-driven proliferation of the HIV-1 proviral reservoir. AIDS Res Ther 14, 7 (2017). https://doi.org/10.1186/s12981-017-0135-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12981-017-0135-1