Abstract
Background
Drug development is currently hampered by high attrition rates; many developed treatments fail during clinical testing. Part of the attrition may be due to low animal-to-human translational success rates; so-called “translational failure”. As far as we know, no systematic overview of published translational success rates exists.
Systematic scoping review
The following research question was examined: “What is the observed range of the animal-to-human translational success (and failure) rates within the currently available empirical evidence?”. We searched PubMed and Embase on 16 October 2017. We included reviews and all other types of “umbrella”-studies of meta-data quantitatively comparing the translational results of studies including at least two species with one being human. We supplemented our database searches with additional strategies. All abstracts and full-text papers were screened by two independent reviewers. Our scoping review comprises 121 references, with various units of measurement: compound or intervention (k = 104), study/experiment (k = 10), and symptom or event (k = 7). Diagnostic statistics corresponded with binary and continuous definitions of successful translation. Binary definitions comprise percentages below twofold error, percentages accurately predicted, and predictive values. Quantitative definitions comprise correlation/regression (r2) and meta-analyses (percentage overlap of 95% confidence intervals). Translational success rates ranged from 0 to 100%.
Conclusion
The wide range of translational success rates observed in our study might indicate that translational success is unpredictable; i.e. it might be unclear upfront if the results of primary animal studies will contribute to translational knowledge. However, the risk of bias of the included studies was high, and much of the included evidence is old, while newer models have become available. Therefore, the reliability of the cumulative evidence from current papers on this topic is insufficient. Further in-depth “umbrella”-studies of translational success rates are still warranted. These are needed to evaluate the probabilistic evidence for predictivity of animal studies for the human situation more reliably, and to determine which factors affect this process.
Similar content being viewed by others
Background
The general aim of biomedical research is to develop possible cures for diseases. Current drug development is handicapped by high attrition rates; many molecules that were promising during preclinical development fail during subsequent clinical testing [1]. At the moment, return on investment in pharma is lower than ever [2]. Part of the attrition may be due to low animal-to-human translational success rates; the so-called “translational failure” [3].
There are two fundamental perspectives potentially explaining translational failures. The first main perspective is that the concept of animal-to-human predictability is fundamentally mistaken [4]. This perspective is based on the observation that the hypothesis that animals are predictable for humans has never been scientifically tested [5, 6], and that there are important differences between species in e.g. physiology, genetics, epigenetics and molecular biology [4, 7]. Animal studies have historically been implemented in drug approval procedures, which may have been based on scientifically outdated principles [8]. Besides, animals and humans are complex systems, that are more than the sum of their parts, and therefore always unpredictable [9, 10]. From this perspective, animal experiments that are performed to inform human health are not ethically acceptable.
The second main perspective is that biomedical and pharmaceutical research advanced over the last decades because animal experiments are in general able to predict the situation in humans [11]. In this perspective, recent translational failure could be explained by suboptimal experimental design [12, 13], and lack of reproducibility in general [1]. Many of the factors involved in suboptimal design of animal studies and the resulting bias, have been reviewed before, and are increasingly taken into account by the scientific community [14,15,16,17,18,19].
Both perspectives are currently promoted by different groups of scientists. Neither group routinely refers to the total body of available evidence on animal-to-human predictability. This predictability, i.e. the translational success rates, can be determined quantitatively in various manners. For example, researchers can sample clinical trials from a registry, retrieve the supporting preclinical data and analyse to what extent the data correspond. Alternatively, they can sample preclinical studies with relevance to humans, and analyse subsequent clinical studies. Moreover, researchers can analyse the effects of a set of interventions (e.g. drugs) on specific outcomes (e.g. biochemistry, physiology and adverse events) in multiple species.
Several methods have been used to analyse translational success, and many authors have addressed the problem of attrition in translational research, e.g. [3, 16, 20]. Most of the papers published on the topic provide expert opinions, narrative reviews or primary studies showing mechanistic similarities between species. As far as we know, no proper overview of the actual data is currently available. While the debate will not be decided by these data alone, an overview of the observed predictive values in different data sets will aid the ethical discourse on the acceptability of those animal experiments intended to inform decisions for human exposure.
In medicine, systematic reviews (SRs) have long been considered to provide the highest level of research evidence, as they combine all available data [21]. In animal research, SRs are increasingly used to collate all available evidence on a subject using transparent and reproducible methodology. We set out to summarise all available published evidence on animal to human translational success. Due to the lack of specific and sensitive indexing of this type of studies, performing a full comprehensive search to retrieve all available studies was not viable. We thus performed a systematic scoping review. Scoping reviews aim to estimate the size and quality of literature on a topic [22]. In the present systematic scoping review, we fully analysed the included papers, to summarise quantitative data from studies that assessed animal-to-human translational success rates.
The main question was “What is the observed range of the animal-to-human translational success (and failure) rates within the currently available empirical evidence?” In contrast to a full systematic review, our review did not follow the PICO format for outcome measures as we included all relevant outcomes, and it did not comprise a full comprehensive systematic search. The search was supplemented by alternative strategies, as detailed in “Methods” section.
Besides studies explicitly addressing translational success rates, we included meta-analyses comprising both human and animal data, and studies analysing the correlation of similar outcomes between animals and humans, as they provide quantitative information on translation for individual interventions. As far as we are aware, we are the first to provide a systematic scoping review of all types of published findings (i.e. literature reviews and other types of “umbrella”-studies) on quantitative analyses of animal-to-human translational success.
Methods
The protocol for this systematic scoping review was posted online on the SYRCLE website (http://www.SYRCLE.nl) on 27 December 2017 [23], after performing the Pubmed and Embase searches, but before the start of paper selection.
Research question
The main research question for this systematic scoping review was: “What is the observed range of the animal-to-human translational success (and failure) rates within the currently available empirical evidence?”. We originally defined translational success as “replication in a randomized trial in humans of statistically significant positive, negative or neutral results for the primary study outcome in animal experiments”, and consequently, translational failure as not replicating the results of animal experiments in a randomized trial for the primary study outcome. We did not expect to find clinical trial publications after animal experiments with negative or neutral efficacy results, nor did we expect to find many after positive toxicology results.
We intended to preferentially address studies on phase I–II trials to focus on early clinical trials over market access, as successful early trials do not always result in clinically available medication for reasons beyond animal-to-human predictability. In practice, only few of the included references detailed the types of trials and experiments, or the primary study outcomes. During the screening of the retrieved references, these two elements thus had to be disregarded. Our working definition of translation therefore became “the quantitative degree of correspondence between the results from a trial in humans with results in animal experiments”. This was communicated between the screeners. We did not post an amendment to the protocol because we did not expect this broader definition to increase bias in the selection of studies and thereby the results.
Search
Our search consisted of 3 elements: animal models, human trials, and translation. We first tested several traditional comprehensive search strategies based on both medical subject headings (MeSH) and on words in the title, abstract and keywords in PubMed. Regardless of the exact combination of search terms used, the number of references retrieved became too high to manage within the timeframe of this project. We thus went for a less conventional scoping strategy, searching for MeSH-terms and title words only. As we expected to miss relevant literature this way, we introduced additional search strategies (detailed below).
Our final search for Pubmed consisted of MeSH-terms and title words (including several synonyms) for animal models, human clinical trials and translation, combined with “AND”. We built an equivalent search for Embase (replacing MeSH terms with the corresponding Emtree terms), also including key words. We filtered for (systematic and other) reviews, letters and editorials. The full search strategy can be found in our protocol [23]. We performed our systematic scoping search in Pubmed and Embase on 16 October 2017.
Additional search strategies
Besides formal literature searches, we retrieved relevant references via two more routes. The first was screening of the reference lists of all included references and relevant reviews. This is a standard approach in systematic reviews. The second alternative route was contacting experts in the field for additional references. Experts were (1) the authors; (2) their (direct and indirect) colleagues known to be interested in translational success, and (3) the first and last authors of all papers included from the search. Experts were contacted via email; a single reminder was sent after 1–2 weeks if they did not respond.
Selection of papers
We included studies and reviews quantitatively comparing the results of studies including at least 2 species with one being human. We thus excluded studies and reviews comparing 2 non-human species or comparing outcomes between human clinical trials. There were no restrictions for language or publication date. All titles and abstracts retrieved from the search were independently screened by two reviewers. Full-text screening of the included papers was again performed by two independent reviewers. Discrepancies were resolved by discussion between reviewers.
During the selection process, we came across several correlational studies of pharmacokinetic–pharmacodynamic (PKPD) parameters after the administration of various molecules. While these papers do not describe translational success rates according to our original binary definition (replication of positive, negative or neutral results), they do provide continuous quantitative information on animal-to-human translation. As this is in line with our intended goal, we did include these papers. The same argumentation led to the inclusion of meta-analyses in which both human and animal studies were compared as subgroups within a single meta-analysis.
Comparisons of outcome measures without an intervention were excluded (e.g. [24,25,26]), as well as papers describing the effects of experimental design parameters on outcomes in several species (e.g. [27]. Ex vivo and in vitro animal studies were also excluded (e.g. [28, 29], as well as animal studies combining the animal with other (mostly in vitro) data to improve predictive accuracy [30]. We only included studies that provided quantitative information on translational success (or failure); i.e. we excluded papers comparing a single human with a single animal study. The unit of analysis could vary, but included studies had to compare a specific set of compounds/interventions, studies/experiments, or symptoms/events between species. The important work of O’Collins et al. [31] was excluded from our analyses as their efficacy comparison between species is not based on the same set of drugs.
Besides, several important reviews focusing on translation from the animal study perspective only were excluded (e.g. [32,33,34,35,36]), as well as studies analysing how often animal studies were cited [37]. Further excluded were important papers on attrition rates and translation with a wider scope than animal-to-human translation (e.g. [3, 38,39,40,41,42,43,44,45,46]), papers presenting relevant graphs without informing us on summary values (e.g. [5], and quantitative studies on related phenomena such as market withdrawal of drugs [47] and animal harm–human benefit analyses [48].
Selection of data
When a single paper described multiple studies on different datasets, those compliant with our inclusion criteria were included into the analyses separately. E.g. [49] described 3 studies of which 2 are included in this review; the 3rd study, on intestinal expression levels of transporters and metabolic enzymes in rats and humans, did not comprise an intervention and was thus excluded.
If species were analysed separately, we included the separate data. If multiple analyses with the same outcome measure were based on the same data, we included the one with the largest sample size (which was deemed the most reliable), or, in a minority of studies with equal sample sizes, the most predictive one (i.e. the highest translational success rate). Including the most predictive results may have biased our results somewhat towards inflated translational success rates. For PKPD studies reporting ≥ 3 parameters, we preferentially selected the volume of distribution, the clearance and the half time, as these were most frequently reported.
For papers describing several meta-analyses based on the same studies, the primary outcomes were selected. If no primary outcome was described, again, the largest analyses were preferentially selected. If multiple binary analyses were based on the same data, we preferentially included the accuracy (see Table 1). If negative and positive predictive value (see Table 1) were both provided without the crude data (from which we could have calculated the accuracy), we included both values.
Analyses of translational success rates
As we observed a large range of reported translational success rates, we exploratively analysed the data further. However, different papers used different strategies for quantifying translation. It is important to realise that different definitions for translational success result in different diagnostic statistics, which may result in different values for the same data.
Different diagnostic statistics lead to different predictive values, even when based on the same data, which we included as described in the preceding section. The differences are clear for e.g. the percentage < twofold error versus the correlation [50], and for the sensitivity, specificity, positive predictive value and negative predictive value [51]. The data are not always so discrepant. For example, the percentage of overall correct predictions reported by Litchfield is 74% when both rat and dog are considered [52]. We can also use his data to calculate specificity (72%), sensitivity (76%), positive predictive value (68%), and negative predictive value (79%).
The diagnostic statistics can be clustered into two main categories: continuous (degree of comparability in effect size; e.g. correlation or % overlap in confidence interval) and binary (translation yes/no; e.g. percentage accurately predicted or percentage below twofold error). For a direct comparison of continuous outcomes, analyses of correlation and regression were common. For yes/no type decisions, several binary classification measures were used, as described in Table 1.
Besides various analyses resulting in different values, different types of values for translational success have different meanings. For example, when analysing percentages of binary success/failure rates, 50% is equivalent to tossing a coin, while 50% overlap in a confidence interval (meta-analyses), or 50% explained variance (correlation and regression) can be considered to result in meaningful data.
If the authors of a paper did not provide a summary measure for translation, we calculated one where we could. For example, for a study on the predictive validity of pain models [53] we calculated the correlation coefficient for the maximum plasma concentration at the minimum effective dose in rats and the maintenance dose in humans using Excel. When different sources were provided describing different values for a single data point (e.g. different values for a single drug in a correlational analysis), we used the median.
All included studies were tabulated. In our tabulations of study outcomes, we aimed to summarise the data and reflect the original authors’ view. To summarise our findings quantitatively, we expressed all values for translation as percentages. The conversions are described in Table 2. For correlations and regression analyses, we selected r2 over r, as this value reflects the percentage of explained variation. When both correlation and % < twofold error were presented, we selected r2 for inclusion in the analyses as these values better reflect the actual data. Similarly, when binary classifications were provided, we preferentially selected accuracy, or, when accuracy was not given, the positive and negative predictive values. For meta-analyses, we determined the degree of overlap of the animal 95% confidence interval (CI) with the human 95% CI. There was only one study where the 95% CIs did not overlap, and in that study, the direction of the effect was opposite in animals compared to humans, included in the analyses as 0% translational success.
To visualise the variation in reported translational success rates, we plotted all values from all included studies in a histogram. We then created boxplots with the individual data points in overlay. Plots were created in R version 3.5.0—“Joy in Playing” [54], using the GGPlot2 package [55].
Risk of bias and reporting quality
According to the protocol, we analysed risk of bias and reporting quality of the included references for the following items: power calculation for the translational comparison, sampling method of the studies included in the analysis, type of data analysis, blinding in the sampling procedure, blinding of the data analyst, control for publication bias (i.e. did the authors analyse the effect of potential underreporting of small neutral studies in their estimate of the translational success outcome), risk of bias analysis performed for each of the included studies, and overall risk of bias estimate. For each of these items, we separately analysed if they were reported, and if there were resulting risks of bias (yes/no/unclear). Of note, power analyses are not common in literature reviews, as systematic reviews aim to include all available evidence, and with complete sampling, power calculations become irrelevant.
Besides, as the included papers described some type of review of the literature, we analysed their compliance with the PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analyses) guidelines for the following items (being aware that these guidelines do not necessarily apply to other review types): registration of a protocol, explicit description of eligibility criteria, the number of screeners determining which papers to include, the number of scientists performing the data-extraction, whether an analysis of risk of bias was performed on the included studies individually and overall, if the analyses had been prespecified, if the limitations of the review were discussed, and if the funding was described. At the time we posted our protocol, the more relevant PRISMA extension for scoping reviews [56] had not yet been published.
For funding, we further estimated whether there was a potential risk of funding bias, which could go either way. We scored a high risk of funding bias if the funder was indicated as or if any of the authors worked for a non-governmental (animal rights) organisation, a pharmaceutical company, or a governmental regulating agency (e.g. EMA, FDA).
Only those relevant PRISMA items not otherwise analysed were extracted. E.g. item 13 and 14 (summary measures and synthesis of results) overlap with our extracted data on type of analysis, and e.g. items 1 and 2 (title and summary) were not deemed relevant for the overview provided in this paper. The item publication bias can be considered irrelevant for certain types of review, for example when internal databases are used. Where included references were not based on publications, we reinterpreted this item for the type of data included, e.g. the risk of studies not ending up in the internal database from which a dataset was extracted.
Results
Search and selection
Our search in PubMed retrieved 2486 references, that in EmBase retrieved 484. After duplicate removal, 2649 references remained, after title-abstract screening, 287. After full-text screening, 26 references were included in this review. Screening the reference lists resulted in 60 additional references. Contacting the first and last authors of the 26 references included from the search combined with contacting people in our network resulted in 35 additional references. The flow of papers is shown in Fig. 1.
Characteristics of the included papers
Of the 121 included references, 119 were in English, one was in German, and one was in French. The unit of measurement was compound or other type of intervention for 104 references, study/experiment for 10, and symptom or event for 7. The number of included interventions, studies or symptoms per reference ranged from 5 to 1256 (also see Fig. 8). Specific animal models were described in 35 references, and comprised e.g. xenografts, bile duct cannulated animals, chimeric mice, or a combination of various models.
Reporting information was limited; less than 15 references reported sex, age or disease status of the animals or humans included in the analyses and the type of studies or trials. Information on dose was reported in 24 references, information on route of administration in 40 (mainly multiple and intravenous, also oral, intraperitoneal and topical). These data were not further analysed.
Studies addressing translational success rates
Studies addressing general medical sciences and efficacy
Of the 121 included references, 16 addressed efficacy or translation in general. The results from these references are summarised in Table 3. Several of these references were familiar to the authors before starting this work and provided the background for our protocol [57,58,59]. Lindl et al. followed the results from 51 animal ethics requests, and found very little translation to the clinical situation [58], with their analysis restricted to a 10-year time window. This may be rather short for analysing translational success, as the development of new treatments is a lengthy process, (see e.g. [39]) and development times seem to increase over time [2]. Hackam et al. followed highly cited animal studies, and found that about one-third translated to randomised clinical trials [57]. Perel et al. compared the effects of 6 interventions between animals and humans with systematic literature reviews, with half of them concordant [59]. We hoped to retrieve a number of comparable references, but only found one; Contopoulos-Ioannidis et al. analysed 101 articles that described novel therapeutic or preventive promises based on animal data [60]. 16 of these novel interventions were tested in clinical trials, of which 12 had a positive result in the trial.
Three references compared the number of positive-outcome studies between animals and humans for similar interventions [61,62,63]. Four other included references comprised meta-analyses showing both human and animal data [64,65,66,67].
The included analyses comprise correlational analyses (R2), the Chi-square test, relative risk, accuracy and meta-analyses.
Studies analysing adverse events and toxicology
Of the 121 included references, 28 addressed translation of safety studies. Adverse events were analysed in 17 of these, carcinogenicity in 6. The other 5 references described translation for drug-induced liver injury, QT prolongation, skin sensitization, teratogenicity and toxic dose. The included references comprise analyses of concordance, likelihood ratios, positive and negative predictive values, sensitivity, Chi-square and correlation. The results from these references are summarised in Table 4.
Of note, the 4 references (comprising 6 studies) with sample sizes over 200 all fall within this category [68,69,70,71]. Tamaki et al. studied 1256 adverse drug reactions after administration of 142 drugs that were approved in Japan from 2001 to 2010 [71]. 48% of the adverse drug reactions could be predicted from the animal data. Fourches et al. mined the literature to create a data set of 951 compounds with effects in the liver in different species [69]. The concordance of liver effects between animals and humans was relatively low. Olson et al. described 221 human toxicity events after administration of 150 (coded) compounds, as reported by 12 pharmaceutical companies [70]. The concordance rates between animal and human toxicity were 71% when all species were considered, with nonrodents alone predictive for 63% and rodents for 43% of the events. Alden et al. reviewed drug labels from the physicians’ desk reference, which they searched for any mention of terms related to carcinogenesis [68]. This resulted in 533 active pharmaceutical ingredients that were further analysed. Of these, 287 had been tested in rodents, in which 161 tested positive for carcinogenicity. The authors presented the sensitivity (73%), positive predictive value (20%, refer to Table 1 for an explanation of predictive values; true positives are in this case ingredients that show carcinogenicity in animals and humans), negative predictive value (90%) and crude data.
Studies addressing pharmacokinetics
Of the 121 included references, 77 addressed translation of various pharmacokinetic (PK) parameters, mainly clearance, bioavailability, volume of distribution and concentration–time profiles. The results from these references are summarised in Table 5. Besides animal–human correlations for PK values from several drugs, these studies often analyse the fold-error of the predicted compared to the observed value, and the percentage of compounds with a predicted value within twofold of the observed value.
Several scatterplots of pharmacokinetic parameters for a set of drugs in animals versus humans show low correspondence rates, i.e. they do not show an apparent relationship between animal and human data (e.g. [72, 73]. Of note, these types of plots are specifically sensitive to selection bias; if one is familiar with the literature it is relatively easy to (consciously or subconsciously) select a set of drugs with relatively high or relatively low correspondence. Besides, PK correlational review papers are often based on the same experiments and data; the same data have e.g. been included in [74,75,76].
Hypotheses-generating analyses of translational success rates
The range of published translational success rates is 0% to 100%. A histogram of all published translational success rates is provided in Fig. 2.
As we included outcomes from different types of analyses, we compared the effect of the two broadly defined definitions of translation; binary (translation successful yes/no) and continuous (amount of correspondence; explained variance) in a boxplot (Fig. 3). For studies using binary definitions of translation, translational success rates ranged from 0 to 93%. Binary definitions comprise the diagnostic statistics fold error (i.e. the percentage of studies/compounds below twofold error), percentage of studies/compounds/adverse events accurately predicted, positive predictive values and negative predictive values. For studies using quantitative definitions of translation, translational success rates ranged from 0 to 100%. Quantitative definitions comprise the diagnostic statistics correlation/regression (r2 expressed as a percentage) and meta-analyses (percentage overlap of 95% confidence intervals of the summary measure). The outcomes of analyses of translational success could be affected by the choice of definition, but the range is large either way.
As we included reviews using different units of analyses, we compared the effect of the unit: event (e.g. a specific adverse event), intervention (mostly drugs) or study (or publication) on translational success rates in a boxplot (Fig. 4). For the 8 studies analysing events, the translational success rate ranged from 7 to 74%. For the other two units of analysis, ranges comprised the full spectrum of 0–100% translational success.
We copied the translational success rates from the authors where possible, but also included papers for which the summary measure of interest was not directly given (e.g. manually calculating a correlation or a percentage overlap in 95% CI, refer to the methods for further information). We visually compared the percentages calculated by us with those calculated by the authors of the included papers in a boxplot (Fig. 5). Both categories comprised the full range of 0–100% translational success.
We then grouped the included studies into broad research categories: toxicology, PKPD and efficacy. Translational success rates by category are shown in Fig. 6. No clear differences are observed between these categories. Differences may still be present between more precisely defined medical fields (e.g. cardiovascular disease, neuroscience, inflammation, oncology), but in-depth analysis of differences in translational success rates between these fields is not possible based on the available data, as most fields have been analysed only once or twice.
We next grouped the included studies by species. Translational success rates by species are shown in Fig. 7. Several references did not specify the species used, several others only presented data from several species pooled. Only few studies were available on guinea pigs, only one on pigs. No clear differences are observed between species.
Our next analysis shows the reported translational success rates by study size (i.e. number of included compounds/interventions, studies/experiments, or symptoms/events, all referred to as K, Fig. 8). The studies with n > 200 are all toxicology studies using a binary definition of translation and have been described above.
To test the potential effects of the various search strategies used, we compared the translational success rates between studies retrieved via our network, reference lists and database searches. Translational success rates by source are shown in Fig. 9. No clear differences are observed between these sources; all ranges comprise translational success rates of 2–99%.
Our last analysis shows the reported translational success rates by publication date (Fig. 10). We observe an increase of both the numbers of studies and the observed range of translational success over time.
Risk of bias and reporting quality
Our analysis of the reporting quality of the included references and the risk of bias is summarised in Fig. 11. Many details of the review designs were poorly reported, resulting in an overall unclear risk of bias for our scoping review.
Reporting of the selected PRISMA-items was also poor; none of the references described the posting of a protocol, the number of people screening the papers, the number of people extracting the data, or prespecifying the analyses. Specific eligibility criteria were described in 31 out of the 121 references (26%), limitations by 37 out of the 121 references (31%).
Out of the 121 references, 27 contained specific information on the funding. Risk of funding bias could work in two directions; studies funded by animal rights organizations are expected to find lower than average translational success rates while those with funding from pharmaceutical companies and governmental organizations may be more inclined to overestimate translational success. If we include the affiliations of the authors in our risk of bias assessment for the funding, 81 out of the 121 references had a high risk of funding bias.
Conclusion
General considerations
This systematic scoping review of reviews provides an overview of research efforts on translational success rates. It shows that the amount of available evidence and the overall quality are limited, and that there is high variability between study types. The published translational success rates range from 0 to 100%. The wide range of translational success rates observed in our study, and the lack of a clear relationship with any of the analysed factors, might indicate that translational success is unpredictable; i.e. it might be unclear upfront if the results of primary studies will contribute to translational knowledge. However, the risk of bias of the included studies was high, and much of the included evidence is older (note that this is a review of reviews, the most recent included reviews will be based on older data), while newer models have become available. Therefore, the cumulative evidence of current papers on this topic is insufficient and further “umbrella”-studies of translational success rates are still warranted.
We included studies on animal-to-human translation. We originally defined successful translation as replication in a randomized trial in humans of statistically significant positive, negative or neutral results for the primary study outcome in animal experiments. However, we did not define “replication”. When writing the protocol, we intended to include studies based on systematic reviews [59], animal ethics requests [58] and highly cited animal publications [57]. The set of included studies however also comprises many correlational and modelling PK studies, in line with our adapted definition of translation: “the quantitative degree of correspondence between the results from a trial in humans with results in animal experiments”.
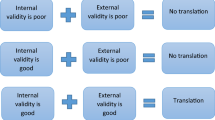
We do not see a difference in predictivity between toxicology, PKPD and efficacy studies. Before we ran the analyses, we expected the toxicology studies to be more predictive than the efficacy studies, first, because toxicology may reflect more conserved physiological mechanisms, second, as toxicology studies are generally performed in multiple species, and third, as toxicology studies are generally performed according to Good Laboratory Practice standards, resulting in higher internal validity of the results.
Search
Scoping searches taught us that a full comprehensive search strategy would result in large numbers of retrieved references, with limited sensitivity. As the resulting amount of work was not manageable within a reasonable time frame, we opted to perform a scoping review instead of a full systematic review, with an in-depth analysis of a subset of the literature.
Our search was thus based on thesaurus (i.e. indexed) and title words only, resulting in missing those papers only describing translation or predictivity in the abstract or the text body while not being indexed for them. We supplemented our search with alternative strategies, i.e. screening the reference lists, contacting first and last authors, and contacting our network, to compensate. We retrieved more references via these alternative strategies (i.e. 60 + 35 = 95, Fig. 1) than via our searches (i.e. 26, Fig. 1), underlining the need for improved indexing of this type of studies.
Snowballing via reference lists is not an optimal method in this field, first because referencing practices are suboptimal (compare e.g. the data and figures from [5, 72, 73, 77]. Second, many studies focussing on alternatives to animal studies also contain information that quantitatively compares animal and human data (e.g. [78] and these relevant studies are difficult to identify from their titles.
A limitation of our search is that we did not include a term for modelling and scaling studies, as we did not have this type of study in mind at the time of designing our protocol. These studies may not specify translation or prediction in their title, e.g. [79, 80]. While using these terms will increase the number of irrelevant hits, to be complete, we do recommend adding the terms “modelling”, “scaling”, “correlation” and their synonyms to retrieve these papers in future searches for translational studies.
A further limitation is that we performed our search halfway October 2017, which is rather inherent to the systematic approach. Systematic reviews of clinical studies take on average 67.3 weeks from registered start to publication [81]. The alternative supplementary strategies increase the review duration, as screening of reference lists and contacting authors of the included studies could only be finalised after full-text screening had been completed and discrepancies between reviewers had been resolved.
We are aware that not all available evidence has been included. To prevent eternal snowballing and to finish this review in a timely manner, we decided to stop retrieving further references from the second-line reference lists onwards. During data-extraction, our occasional checks of reference lists of the later included papers showed that most studies had already been included, indicating that, for a scoping review, our data-set can be considered as almost complete.
A full systematic review following the methodology described in this scoping review would probably result in a larger data set. However, we cannot envision our alternative search strategies to be biased towards a certain outcome. Contacting the authors of the papers retrieved by the search is relatively objective and reproducible. The authors’ network should not induce substantial bias either, as the opinions on translational success rates between the authors vary. The main outcome of this study is the observed range of translational success rates. As this comprises all possible values (0–100%) it could not change because of more complete sampling strategies.
Data quality and risk of bias
Some of the general issues with analysing translational failure and success rates have been described before [82]. Besides, our analyses are affected by several factors. Factors generally affecting the quality of scoping reviews comprise publication bias, unblinded data selection, unblinded extraction, unblinded analysis and statistical power. Publication bias is the relative underreporting of studies not showing a significant effect in scientific literature. For the observed range of reported translational success rates, from 0 to 100%, we do not consider publication bias a specific concern. We strove to limit bias in the inclusion of data by having two reviewers select papers independently. Of note, the observed range of translational success was not drastically affected by publication date or manner of publication retrieval. Data extraction and analysis were not performed in a blinded manner, but the extractor (CHCL) had no a priori expectations on the results. As data were not quantitatively analysed, statistical power is not relevant.
Besides, several factors specifically affect the quality of the data included in this work. The first is the problem with dependency of the data; several authors and research groups publish multiple papers on translational success rates, often based on (partially) overlapping data sets. For example, Schein present an analysis of 25 anticancer drug toxicities in several papers [83,84,85,86], each paper combining the analysis with other information. For our quantitative analyses, we aimed for incorporating each dataset only once, but if datasets only overlapped partially, they were both included.
The second is that we included several measures for translational success, based on different definitions, using different diagnostic statistics. We classified the different measures into two broad categories, based on the underlying definition of translational success, which could be binary (yes/no) or continuous (% concordance), and did not observe large differences in observed translational success rates between these categories. Translational success rates were also not affected by unit of measurement (event, intervention or study) used in the original review, or by who did the calculations (us or the authors of the review). However, the percentage overlap in CIs, which we used to describe translational success for meta-analyses, is disputable for two reasons. First, the overlap in CIs could be fair even if the estimates are quite far apart if both estimates are unprecise. We consider the CIs of the included studies not to be that large. Second, many scientists argue that the size of the effect is irrelevant as long as the direction of the effect is the same. As described in our methods, for the included meta-analyses, only one set of CIs, from rats and humans, did not overlap [65]. The direction of the effect here was opposite, and we included it in our analyses as 0% concordance. We preferred including the percentage overlap over excluding the meta-analyses from our review, and excluding this paper would not have affected the overall observed range of translational success.
A third factor is that dosing and incidence of events are often disregarded [82]. Concerning dosing, differences in metabolism, weight, distribution volume etc. result in different dosing, and oversimplified approaches for dose prediction are common [87]. Concerning incidence, known human carcinogens may be tested more extensively in animals than compounds without known human toxicity, eventually showing positive results in at least one preclinical test. Besides these factors, publication bias (i.e. the relative underreporting of primary studies with negative and neutral results) can obscure translational failure rates [13].
Our analysis of risk of bias in the included references shows an overall unclear risk of bias, with a high risk of funding bias for 81 out of the 122 included references. Besides, there was a high risk for underpowered studies in 30 out of 121 included references. Our analysis of the reporting quality of the included studies showed that most reviews did not comply with the PRISMA guidelines, but this is not unexpected, as most of the included references did not claim to be systematic reviews.
Many of the included reviews had drugs as the experimental unit. Most of these did not describe their selection of the drugs, explicit inclusion and exclusion criteria were scarce. One that did transparently describe their selection procedure excluded studies with novel targets, where predictivity is most needed [88]. This same study shows that analysing a subset excluding outliers can increase the predictivity of the animal studies.
To conclude, the data presented in this paper have severe limitations. They should be considered inconclusive and used for hypothesis-generation only. Besides, reliably determining actual translational success rates is unmanageable as long as the current status of reporting of preclinical research leaves room for improvement [89], and non-reproducibility is such a critical issue in both animal [1] and human [90] studies.
Implications
While the quantity and quality of the available data is limited and further studies are still needed, this review provides an at least relatively complete overview of published evidence on translational success rates. These actual numbers for predictiveness are theoretically more informative than qualitative, subjectively determined, mechanistic similarities between animal models and human pathology. Therefore, for animal studies aimed at translation to the human situation, where possible, probabilistic evidence for predictivity should be considered besides or even instead of mechanistic evidence.
Of note, animal studies may contribute to successful translation in other manners than direct prediction of the human response; they can for example be informative in hypothesis-generation for mechanisms underlying disease. We emphasise that this review does not provide any information on the usefulness of animals in fields of animal use that do not directly target predictivity for humans.
To ensure validity of the gathered animal and human data, it is essential that the execution of the studies is of high quality, and that the reporting is complete. Complete reports of high-quality studies are needed to determine actual translational success rates, and to identify factors involved in translational success. Knowing the factors involved in translational success will benefit both animals and humans.
Availability of data and materials
All included data are available in the public domain and all references are included in our reference list. Extracted data and calculations will be made available to individual scientists upon reasonable request.
Abbreviations
- AN:
-
animal negative
- AP:
-
animal positive
- CI:
-
confidence interval
- FN:
-
false negative
- FP:
-
false positive
- HN:
-
human negative
- HP:
-
human positive
- LR:
-
likelihood ratio
- MeSH:
-
medical subject heading
- NPV:
-
negative predictive value
- PK:
-
pharmacokinetic
- PKPD:
-
pharmacokinetic–pharmacodynamic
- PPV:
-
positive predictive value
- PRISMA:
-
preferred reporting items for systematic reviews and meta-analyses
- SR:
-
systematic review
- TN:
-
true negative
- TP:
-
true positive
References
Freedman LP, Cockburn IM, Simcoe TS. The economics of reproducibility in preclinical research. PLoS Biol. 2015;13(6):e1002165.
Steedman M, Taylor K, Stockbridge M, Korba C, DShah S, Thaxter M. Unlocking R&D productivity—measuring the return from pharmaceutical innovation 2018. 2019.
Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3(8):711–5.
Pound P, Ritskes-Hoitinga M. Is it possible to overcome issues of external validity in preclinical animal research? Why most animal models are bound to fail. J Transl Med. 2018;16(1):304.
Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med. 2009;4:2.
Shanks N, Greek CR. Animal model in light of evolution. Boca Raton: BrownWalker Press; 2009.
Rust JH. Animal models for human diseases. Perspect Biol Med. 1982;25(4):662–72.
Greek R, Pippus A, Hansen LA. The Nuremberg code subverts human health and safety by requiring animal modeling. BMC Med Ethics. 2012;13:16.
Greek R, Hansen LA. Questions regarding the predictive value of one evolved complex adaptive system for a second: exemplified by the SOD1 mouse. Prog Biophys Mol Biol. 2013;113(2):231–53.
Greek R, Shanks N. Complex systems, evolution, and animal models. Stud Hist Philos Biol Biomed Sci. 2011;42(4):542–4.
Degeling C, Johnson J. Evaluating animal models: some taxonomic worries. J Med Philos. 2013;38(2):91–106.
Bolker JA. Animal models in translational research: rosetta stone or stumbling block? Bioessays. 2017;39(12):1700089.
Martic-Kehl MI, Schibli R, Schubiger PA. Can animal data predict human outcome? Problems and pitfalls of translational animal research. Eur J Nucl Med Mol Imaging. 2012;39(9):1492–6.
Garner JP. The significance of meaning: why do over 90% of behavioral neuroscience results fail to translate to humans, and what can we do to fix it? ILAR J. 2014;55(3):438–56.
Garner JP, Gaskill BN, Weber EM, Ahloy-Dallaire J, Pritchett-Corning KR. Introducing therioepistemology: the study of how knowledge is gained from animal research. Lab Anim. 2017;46(4):103–13.
Hartung T. Look back in anger—what clinical studies tell us about preclinical work. Altex. 2013;30(3):275–91.
Green SB. Can animal data translate to innovations necessary for a new era of patient-centred and individualised healthcare? Bias in preclinical animal research. BMC Med Ethics. 2015;16:53.
Smith AJ, Clutton RE, Lilley E, Hansen KEA, Brattelid T. PREPARE: guidelines for planning animal research and testing. Lab Anim. 2018;52(2):135–41.
Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8(6):e1000412.
Duyk G. Attrition and translation. Science. 2003;302(5645):603–5.
de Vries RB, Wever KE, Avey MT, Stephens ML, Sena ES, Leenaars M. The usefulness of systematic reviews of animal experiments for the design of preclinical and clinical studies. ILAR J. 2014;55(3):427–37.
Booth A, Papaioannou D, Sutton A. Systematic approaches to a successful literature review. London: Sage; 2012. p. 279.
Menon JM, Kouwenaar C, Stafleu F, De Vries RBM, Ritskes-Hoitinga, Meijboom FLB, et al. Quantification of translational success: rates of concordance between the results of animal experiments and human trials—a systematized review [protocol posted online on http://www.SYRCLE.nl]. Nijmegen: SYRCLE; 2017.
Dalgaard L. Comparison of minipig, dog, monkey and human drug metabolism and disposition. J Pharmacol Toxicol Methods. 2015;74:80–92.
Oerton E, Bender A. Concordance analysis of microarray studies identifies representative gene expression changes in Parkinson’s disease: a comparison of 33 human and animal studies. BMC Neurol. 2017;17(1):58.
Workman AD, Charvet CJ, Clancy B, Darlington RB, Finlay BL. Modeling transformations of neurodevelopmental sequences across mammalian species. J Neurosci. 2013;33(17):7368–83.
Boyes WK. Rat and human sensory evoked potentials and the predictability of human neurotoxicity from rat data. Neurotoxicology. 1994;15(3):569–78.
Lennernas H, Nylander S, Ungell AL. Jejunal permeability: a comparison between the using chamber technique and the single-pass perfusion in humans. Pharm Res. 1997;14(5):667–71.
Kamimura H, Nakada N, Suzuki K, Mera A, Souda K, Murakami Y, et al. Assessment of chimeric mice with humanized liver as a tool for predicting circulating human metabolites. Drug Metab Pharmacokinet. 2010;25(3):223–35.
Zuegge J, Schneider G, Coassolo P, Lave T. Prediction of hepatic metabolic clearance: comparison and assessment of prediction models. Clin Pharmacokinet. 2001;40(7):553–63.
O’Collins VE, Macleod MR, Donnan GA, Horky LL, van der Worp BH, Howells DW. 1,026 experimental treatments in acute stroke. Ann Neurol. 2006;59(3):467–77.
Amarasingh S, Macleod MR, Whittle IR. What is the translational efficacy of chemotherapeutic drug research in neuro-oncology? A systematic review and meta-analysis of the efficacy of BCNU and CCNU in animal models of glioma. J Neurooncol. 2009;91(2):117–25.
Moeller A, Ask K, Warburton D, Gauldie J, Kolb M. The bleomycin animal model: a useful tool to investigate treatment options for idiopathic pulmonary fibrosis? Int J Biochem Cell Biol. 2008;40(3):362–82.
Theunissen PT, Beken S, Beyer B, Breslin WJ, Cappon GD, Chen CL, et al. Comparing rat and rabbit embryo-fetal developmental toxicity data for 379 pharmaceuticals: on systemic dose and developmental effects. Crit Rev Toxicol. 2017;47(5):402–14.
Theunissen PT, Beken S, Beyer BK, Breslin WJ, Cappon GD, Chen CL, et al. Comparison of rat and rabbit embryo-fetal developmental toxicity data for 379 pharmaceuticals: on the nature and severity of developmental effects. Crit Rev Toxicol. 2016;46(10):900–10.
Woutersen RA, Soffers AE, Kroese ED, Krul CA, van der Laan JW, van Benthem J, et al. Prediction of carcinogenic potential of chemicals using repeated-dose (13-week) toxicity data. Regul Toxicol Pharmacol. 2016;81:242–9.
Dagg AI, Seidle TK. Levels of citation of nonhuman animal studies conducted at a Canadian research hospital. J Appl Anim Welf Sci. 2004;7(3):205–13.
Ciesielski TH, Aldrich MC, Marsit CJ, Hiatt RA, Williams SM. Transdisciplinary approaches enhance the production of translational knowledge. Transl Res. 2017;182:123–34.
Contopoulos-Ioannidis DG, Alexiou GA, Gouvias TC, Ioannidis JP. Medicine. Life cycle of translational research for medical interventions. Science. 2008;321(5894):1298–9.
Cohrs JR, Martin T, Ghahramani P, Bidaut L, Higgins PJ, Shahzad A. Translational medicine definition by the European society for translational medicine. New Horiz Transl Med. 2015;2:86–8.
Davis MM, Butchart AT, Wheeler JR, Coleman MS, Singer DC, Freed GL. Failure-to-success ratios, transition probabilities and phase lengths for prophylactic vaccines versus other pharmaceuticals in the development pipeline. Vaccine. 2011;29(51):9414–6.
Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32(1):40–51.
Prentis RA, Walker SR. Trends in the development of new medicines by UK-owned pharmaceutical companies (1964–1980). Br J Clin Pharmacol. 1986;21(4):437–43.
Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nat Rev Drug Discov. 2011;10(9):712.
Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11(3):191–200.
Waring MJ, Arrowsmith J, Leach AR, Leeson PD, Mandrell S, Owen RM, et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat Rev Drug Discov. 2015;14(7):475–86.
Onakpoya IJ, Heneghan CJ, Aronson JK. Worldwide withdrawal of medicinal products because of adverse drug reactions: a systematic review and analysis. Crit Rev Toxicol. 2016;46(6):477–89.
Pound P, Nicol CJ. Retrospective harm benefit analysis of pre-clinical animal research for six treatment interventions. PLoS ONE. 2018;13(3):e0193758.
Cao X, Gibbs ST, Fang L, Miller HA, Landowski CP, Shin HC, et al. Why is it challenging to predict intestinal drug absorption and oral bioavailability in human using rat model. Pharm Res. 2006;23(8):1675–86.
Caldwell GW, Masucci JA, Yan Z, Hageman W. Allometric scaling of pharmacokinetic parameters in drug discovery: can human CL, Vss and t1/2 be predicted from in vivo rat data? Eur J Drug Metab Pharmacokinet. 2004;29(2):133–43.
Monticello TM, Jones TW, Dambach DM, Potter DM, Bolt MW, Liu M, et al. Current nonclinical testing paradigm enables safe entry to First-In-Human clinical trials: the IQ consortium nonclinical to clinical translational database. Toxicol Appl Pharmacol. 2017;334:100–9.
Litchfield JT Jr. Symposium on clinical drug evaluation and human pharmacology—Part XVI. Evaluation of the safety of new drugs by means of tests in animals. Clin Pharm Ther. 1961;3(5):665–72.
Whiteside GT, Adedoyin A, Leventhal L. Predictive validity of animal pain models? A comparison of the pharmacokinetic–pharmacodynamic relationship for pain drugs in rats and humans. Neuropharmacology. 2008;54(5):767–75.
R_Core_Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2016. https://www.R-project.org/. Accessed 29 Oct 2016.
Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2016.
Tricco AC, Lillie E, Zarin W, O’Brien KK, Colquhoun H, Levac D, et al. PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med. 2018;169(7):467–73.
Hackam DG, Redelmeier DA. Translation of research evidence from animals to humans. JAMA. 2006;296(14):1731–2.
Lindl T, Völkel M, Kolar R. Animal experiments in biomedical research. An evaluation of the clinical relevance of approved animal experimental projects: no evident implementation in human medicine within 10 years. Altern Lab Anim. 2005;22(3):143–51.
Perel P, Roberts I, Sena E, Wheble P, Briscoe C, Sandercock P, et al. Comparison of treatment effects between animal experiments and clinical trials: systematic review. BMJ. 2007;334(7586):197.
Contopoulos-Ioannidis DG, Ntzani E, Ioannidis JP. Translation of highly promising basic science research into clinical applications. Am J Med. 2003;114(6):477–84.
Brossi PM, Moreira JJ, Machado TS, Baccarin RY. Platelet-rich plasma in orthopedic therapy: a comparative systematic review of clinical and experimental data in equine and human musculoskeletal lesions. BMC Vet Res. 2015;11:98.
Steinberg WM, Schlesselman SE. Treatment of acute pancreatitis. Comparison of animal and human studies. Gastroenterology. 1987;93(6):1420–7.
Yardley MM, Ray LA. Medications development for the treatment of alcohol use disorder: insights into the predictive value of animal and human laboratory models. Addict Biol. 2017;22(3):581–615.
Faggion CM Jr, Chambrone L, Gondim V, Schmitter M, Tu YK. Comparison of the effects of treatment of peri-implant infection in animal and human studies: systematic review and meta-analysis. Clin Oral Implants Res. 2010;21(2):137–47.
Sultan SR, Millar SA, England TJ, O’Sullivan SE. A systematic review and meta-analysis of the haemodynamic effects of cannabidiol. Front Pharmacol. 2017;8:81.
Valles C, Rodriguez-Ciurana X, Clementini M, Baglivo M, Paniagua B, Nart J. Influence of subcrestal implant placement compared with equicrestal position on the peri-implant hard and soft tissues around platform-switched implants: a systematic review and meta-analysis. Clin Oral Investig. 2018;22(2):555–70.
Yen CC, Tu YK, Chen TH, Lu HK. Comparison of treatment effects of guided tissue regeneration on infrabony lesions between animal and human studies: a systematic review and meta-analysis. J Periodontal Res. 2014;49(4):415–24.
Alden CL, Lynn A, Bourdeau A, Morton D, Sistare FD, Kadambi VJ, et al. A critical review of the effectiveness of rodent pharmaceutical carcinogenesis testing in predicting for human risk. Vet Pathol. 2011;48(3):772–84.
Fourches D, Barnes JC, Day NC, Bradley P, Reed JZ, Tropsha A. Cheminformatics analysis of assertions mined from literature that describe drug-induced liver injury in different species. Chem Res Toxicol. 2010;23(1):171–83.
Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, et al. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol. 2000;32(1):56–67.
Tamaki C, Nagayama T, Hashiba M, Fujiyoshi M, Hizue M, Kodaira H, et al. Potentials and limitations of nonclinical safety assessment for predicting clinical adverse drug reactions: correlation analysis of 142 approved drugs in Japan. J Toxicol Sci. 2013;38(4):581–98.
Grass GM, Sinko PJ. Physiologically-based pharmacokinetic simulation modelling. Adv Drug Deliv Rev. 2002;54(3):433–51.
Musther H, Olivares-Morales A, Hatley OJ, Liu B, Rostami Hodjegan A. Animal versus human oral drug bioavailability: do they correlate? Eur J Pharm Sci. 2014;57:280–91.
Evans CA, Jolivette LJ, Nagilla R, Ward KW. Extrapolation of preclinical pharmacokinetics and molecular feature analysis of “discovery-like” molecules to predict human pharmacokinetics. Drug Metab Dispos. 2006;34(7):1255–65.
Ward KW, Smith BR. A comprehensive quantitative and qualitative evaluation of extrapolation of intravenous pharmacokinetic parameters from rat, dog, and monkey to humans. II. Volume of distribution and mean residence time. Drug Metab Dispos. 2004;32(6):612–9.
Ward KW, Smith BR. A comprehensive quantitative and qualitative evaluation of extrapolation of intravenous pharmacokinetic parameters from rat, dog, and monkey to humans. I. Clearance. Drug Metab Dispos. 2004;32(6):603–11.
Sietsema WK. The absolute oral bioavailability of selected drugs. Int J Clin Pharmacol Ther Toxicol. 1989;27(4):179–211.
Hoffmann S, Kleinstreuer N, Alepee N, Allen D, Api AM, Ashikaga T, et al. Non-animal methods to predict skin sensitization (I): the Cosmetics Europe database. Crit Rev Toxicol. 2018;48(5):344–58.
Bachmann K, Pardoe D, White D. Scaling basic toxicokinetic parameters from rat to man. Environ Health Perspect. 1996;104(4):400–7.
Chiou WL, Jeong HJ, Chung SM, Wu TC. Evaluation of using dog as an animal model to study the fraction of oral dose absorbed of 43 drugs in humans. Pharm Res. 2000;17(2):135–40.
Borah R, Brown AW, Capers PL, Kaiser KA. Analysis of the time and workers needed to conduct systematic reviews of medical interventions using data from the PROSPERO registry. BMJ Open. 2017;7(2):e012545.
Brooker P. The use of second species in toxicology testing. Altern Lab Anim. 2014;42(2):147–9.
Schein P. The prediction of clinical toxicities of anticancer drugs. Pharmacological basis of cancer chemotherapy. Baltimore: Williams and Wilkins; 1975. p. 383–95.
Schein P, Anderson T. The efficacy of animal studies in predicting clinical toxicity of cancer chemotherapeutic drugs. Int J Clin Pharmacol. 1973;8(3):228–38.
Schein P, Davis RD, Cooney DA, editors. Qualitative aspects of drug toxicity in prediction from laboratory animals to man. In: 5th international congress on pharmacology. 1973.
Schein PS, Davis RD, Carter S, Newman J, Schein DR, Rall DP. The evaluation of anticancer drugs in dogs and monkeys for the prediction of qualitative toxicities in man. Clin Pharmacol Ther. 1970;11(1):3–40.
Blanchard OL, Smoliga JM. Translating dosages from animal models to human clinical trials—revisiting body surface area scaling. FASEB. 2015;29:1629–34.
Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res. 2003;9(11):4227–39.
Pound P, Bracken MB. Is animal research sufficiently evidence based to be a cornerstone of biomedical research? BMJ. 2014;348:g3387.
Ioannidis JP. Contradicted and initially stronger effects in highly cited clinical research. JAMA. 2005;294(2):218–28.
Acknowledgements
The authors would like to thank Julia Menon for assistance in the early review phases, Alice Tillema (Radboudumc) for help in optimising the search strategy, and the Utrecht University R data café for help in writing the code for the analyses in R.
Besides, we are very grateful for the following scientists responding to our request for relevant papers: Peter Bieck, Ulrich Dirnagl, Urban Fagerholm, Daniel Hackam, He Handan, David Howells, Jonathan Kimmelman, Manoj Lalu, Judith van Luijk, Italo Pogessi, Pandora Pound, Philip Schein, Emily Sena, William Sietsema, Chihiro Tamaki, Keith Ward, Kim Wever, Harvey Wong, Hanno Würbel and several others who chose not to be specifically acknowledged.
Funding
ZonMW (40-42600-98-417), NWO (313-99-310) and the Federal State of Lower Saxony (R2N) provided funding for this project.
Author information
Authors and Affiliations
Contributions
CK, FS, RdV, MRH, FM and CL developed the protocol. CK, FS and CL performed the searches and selected the papers. CL extracted the data, FS performed a quality control. RdV, AB and CL analysed and interpreted the data. CL wrote the manuscript. All authors contributed ideas and critically revised several drafts of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not relevant.
Consent for publication
All scientists named in the acknowledgements section below consented to being acknowledged in this publication. We did not include any other individual person’s data in any form.
Competing interests
Merel Ritskes-Hoitinga MR-H is a member of the council of management of the UK registered company/charity Laboratory Animals Ltd (LAL). LAL issues the journal Laboratory Animals. The position is unpaid but travel to LAL meetings is reimbursed. The journal’s profits are used for charitable purposes, subsidising educational projects in laboratory animal science and welfare. The other authors have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Leenaars, C.H.C., Kouwenaar, C., Stafleu, F.R. et al. Animal to human translation: a systematic scoping review of reported concordance rates. J Transl Med 17, 223 (2019). https://doi.org/10.1186/s12967-019-1976-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-019-1976-2