Abstract
Backgrounds
Pseudoprogression disease (PsPD) is commonly observed during glioblastoma (GBM) follow-up after adjuvant therapy. Because it is difficult to differentiate PsPD from true early progression of GBM, we have used a quantitative proteomics strategy to identify molecular signatures and develop predictive markers of PsPD.
Results
An initial screening of three PsPD and three GBM patients was performed, and from which 530 proteins with significant fold changes were identified. By conducting biological functional analysis of these proteins, we found evidence that the protein synthesis network and the cellular growth and proliferation network were most significantly affected. Moreover, six of the proteins (HNRNPK, ELAVL1, CDH2, FBLN1, CALU and FGB) involved in the two networks were validated (n = 18) in the same six samples and in twelve additional samples using immunohistochemistry methods and the western blot analysis. The receiver operating characteristic (ROC) curve analysis in distinguishing PsPD patients from GBM patients yielded an area under curve (AUC) value of 0.90 (95% confidence interval (CI), 0.662-0.9880) for CDH2 and.0.92 (95% CI, 0.696-0.995) for CDH2 combined with ELAVL1.
Conclusions
The results of the present study both revealed the biological signatures of PsPD from a proteomics perspective and indicated that CDH2 alone or combined with ELAVL1 could be potential biomarkers with high accuracy in the diagnosis of PsPD.
Similar content being viewed by others
Introduction
Glioblastoma (GBM) is one of the most malignant brain tumors. After the postoperative use of radiotherapy for GBM became common, a phenomenon termed pseudoprogression disease (PsPD) was identified [1,2]. With the widely implementation of the Stupp protocol for treating GBM, this phenomenon has been inceasingly reported, with an incidence rate varies among reports (5.5%-64%) [3-6]. PsPD is often misdiagnosed as tumor recurrence and misleads the clinical treatment. However, little is known about why PsPD occurs in a subset of GBM patients and the fundamental biological features of PsPD remain unclear [5,7-10].
From a diagnostic perspective, no single imaging technique, including T1-weighted magnetic resonance imaging (MRI), magnetic resonance spectroscopy (MRS), relative cerebral blood volume (rCBV)-based parametric response mapping and 18fluorodeoxyglucose (18 F-FDG)-positron emission computed tomography (PET), has been adequate for differentiating PsPD from true early tumor progression with high sensitivity and specificity [4,5,11-16]. Moreover, molecular biological studies have failed to uncover biomarkers linked to PsPD for clinical use. Although a multitude of genetic and molecular changes involved in GBM, including O6-methylguanine–DNA methyltransferase (MGMT) promoter methylation, isocitrate dehydrogenase 1 (IDH1) mutation, p53 mutation and Ki-67 expression, have been found to be associated with PsPD, the predictive value of these biomarkers remains debatable [5,8,17-19]. Therefore, except for cases of pathological verification, PsPD is still predominantly diagnosed retrospectively. Thus, there is an urgent need for the exploration of more reliable biochemical markers that can accurately identify PsPD.
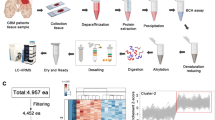
Proteomic measurements provide a wealth of biological information and several proteomic studies of gliomas have been recently reported [20,21], which demonstrated a possibility to investigate this phenomenon by using proteomics methods. Herein, this present study was designed to identify biological signatures and explore biomarkers for PsPD using differential proteomic techniques (Figure 1).
Workflow of the iTRAQ proteomic strategy. In this work, three pathologically verified tissue samples of PsPD and three samples of GBM were used for iTRAQ labeled proteomic analysis. The proteins identified were quantitatively analyzed using Panther and IPA for biological functions analysis. Several candidate proteins with interesting biological functions were selected and further validated using IHC and WB of the same samples used for proteomic analysis as well as additional samples.
Results
Identification of proteins with significant fold changes in PsPD versus GBM
In this iTRAQ-labeling proteomic study, by comparing the total proteomes of tissue from PsPDs with the proteomes of tissues from GBMs, we identified 4048 proteins in PsPD and 3846 proteins in GBM (Additional file 1: File s1, Additional file 2: File s2, Additional file 3: File s3 and Additional file 4: File s4). To measure the quantitative correlation between pairwise sample combinations within each group, a Pearson’s correlation coefficient (ranged from 0.967 to 0.980) was calculated and showed high biological reproducibility (Additional file 5: Figure s1). To maintain a low false-positive rate of comparative analysis between the groups, an average CV of 0.37 (Additional file 5: Figure s2) was employed to filter out data with poor linearity, corresponding to coverage of more than 80% of the 3390 quantified proteins both in PsPDs and GBMs. Next, a threshold of ≥2-fold and p < 0.05 was taken to identify 530 proteins with significant fold changes for further analysis (Figure 2). Among these proteins, 57 proteins were up-regulated and 473 were down-regulated in PsPD compared with GBMs (Additional file 6: File s5 and Additional file 7: File s6).
Interaction networks and functional pathway analysis
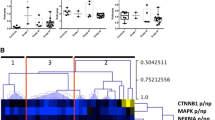
Functional pathway analysis was performed for the 530 proteins to better understand the biological features of PsPD. Gene ontology analysis indicated broad distribution of these proteins, with the most frequently represented categories being cellular compartment, molecular function, and biological processes (Figure 3). The results of Ingenuity Pathway Analysis (IPA) analysis indicated that the protein synthesis network and the cellular growth and proliferation network were mostly affected (Figure 4),with a series of cellular functions being significantly inhibited in PsPD compared with GBM (Additional file 5: Figure s3). For example, the invasion (z-score:-2.575) and proliferation (z-score:-2.886) abilities of tumor cells were significantly downregulated in PsPD compared with GBM. Moreover, the translation (z-score: −2.464), synthesis of protein (z-score: −2.236) and metabolism of protein (z-score:-2.046) were also significantly downregulated in PsPD compared with GBM.
Cellular growth and proliferation network and protein synthesis network from IPA analysis. Graph A shows the protein synthesis focused network, and Graph B illustratescellular growth and proliferation focused function network. Proteins in red were up-regulated in PsPD compared with N-GBM, and proteins in green were down-regulated in PsPD compared with N-GBM. Proteins pointed by the blue arrow were the selected out candidate proteins used for analysis and further validation.
Selection of candidate proteins for validation
Three candidate proteins (HNRNPK, ELAVL1 and CDH2) involved in the two networks and acting as key-point proteins were selected out. In order to explore more promising biomarkers, all secreted proteins with more than 2 fold changes (Additional file 8: File s7) were searched against the protein atlas database (http://www.proteinatlas.org), because the protein atlas database provided the expression levels of candidate proteins in specific tissues and related antibodies. Proteins with median or high positive expression in glial cell or tissue were chosen for further functional analysis. Three proteins (FBLN1, CALU and FGB) meeting the criteria were selected out. The results of IHC and WB validation of the six proteins were in accordance with the proteomic findings (Figures 5, 6). Moreover, a quantitative analysis of the WB results was performed (Table 1, Figure 6). As shown in the figure, statistically significant differences were found between the groups.
Results of immunohistochemical analysis of CDH2, ELAVL1, HNRNPK, FBLN1, CALU and FGB in tissue samples. Magnification: 200X. Representative images of paraffin-embedded sections of PsPD and GBM tissue that were HE stained and immunostained for CDH2,ELAVL1, HNRNPK, FBLN1, CALU and FGB. Graph A shows the validation of these six candidate proteins in the six samples used for proteomic analysis. The first three columns show the validation results in N-GBMs and the second three columns show the results in the PsPDs. Graph B shows the validation in an additional twelve samples. The first four column shows the validation results in additionally selected N-GBMs, the second four column shows the results in R-GBMs, and the third four column shows the results in PsPDs. * indicates the twelve additionally selected samples.
Western blot analysis for ELAVL1, HNRNPK, CDH2 and FBLN1 in tissue samples. Graph A shows that high levels of ELAVL1, HNRNPK, CDH2 and low levels FBLN1 were detected in N-GBMs compared with PsPDs in the six samples for proteomic analysis. Graph B shows the quantification of expression levels using densitometry. Graph C shows that high levels of ELAVL1, HNRNPK, CDH2 and low levels of FBLN1 were detected in GBMs (both N-GBM and R-GBM) compared with PsPDs in additional twelve samples. Graph D shows the quantification of expression levels using densitometry. * indicates the twelve additionally selected samples;** p < 0.05.
Evaluation of HNRNPK, ELAVL1, CDH2 and FBLN1 as diagnostic markers for PsPD
The WB analysis revealed that HNRNPK, ELAVL1, CDH2 and FBLN1 were of statistical significance and exihibited obvious fold changes between PsPDs and GBMs (Table 1). Furthermore, the area under the ROC curves for ELAVL1, HNRNPK, CDH2 and FBLN1 were 0.86 (p = 0.013), 0.75 (p = 0.077), 0.90 (p = 0.006) and 0.66 (p = 0.258), respectively (Figure 7, Additional file 9: Table S1). A pairwise comparison of ROC curves shows no statistical difference between these four proteins (Additional file 9: Table S2). Furthermore, the area under the combined ROC curve for CDH2 and ELAVL1 was 0.92 (P = 0.003), indicating that the diagnostic value of CDH2 alone or combined with ELAVL1 was improved.
Discussion
By using iTRAQ-labeled proteomic analysis and conducting further biological functional analysis of fold-changed proteins, we identified the biological features of PsPD from the perspective of proteomics and explored several candidate proteins to be predictive biomarkers.
Protein metabolism and upstream regulatory mechanisms play fundamental roles
The results of the biological analysis revealed the protein synthesis network to be broadly affected. Based on the data from the present study, the expression level of proteins involved in protein synthesis and upstream regulatory mechanisms, such as RNA post-transcriptional modification, post-translational modification and protein folding are significantly different between PsPDs and GBMs (Figure 4, Additional file 5: Figure s3, Additional file 7: File s5 and Additional file 8: File s6), indicating these mechanisms may be significantly affected. Two candidate proteins, HNRNPK and ELAVL1, involved in the protein synthesis network were selected and validated.
HnRNPs comprise a large family of proteins with approximately 30 members that share some structural domains. Previous studies have shown that hnRNPs played central roles in several cellular functions, among which HNRNPK was found to play an essential role in cellular proliferation by regulating protein synthesis and is over-expressed in head and neck tumors [22,23]. In recent studies, HNRNPK was also found to play a significant role in the mechanism of DNA damage-related cell cycle arrest under ionizing conditions [24,25], which is similar to the effect of radiotherapy. In the present study, hnRNPs (HNRNPC, HNRNPK, HNRNPM and HNRNP) were found to play roles in the protein synthesis network and were down-regulated in PsPDs compared with GBMs, which may reflect the effect of chemo-radiotherapy treatment in GBM patients.
In addition to the hnRNPs, another RNA-binding protein, ELAVL1, was selected. Under hypoxia, ELAVL1 plays a significant role in the regulation of angiogenesis by stabilizing vascular endothelial growth factor A (VEGF-A) mRNA [26,27]. VEGF-A is one of the major mediators of vascular proliferation in astrocytic tumor [28]. Both VEGF and ELAVL1 were identified down-regulated in PsPD compared with GBM, suggesting the possibility of angiogenesis inhibition in PsPD. This result may also help explain how hypoxia is involved in the formation of PsPD, as has been proposed in several studies [18,29].
Cellular function interference
Many researchers have proposed that PsPD occurs due to the induction of cell death by radiotherapy and/or chemotherapy of malignant glioma [17,30]. These findings indicate a hypothesis that an underlying relationship between PsPD occurrence and cell death induction by adjuvant therapy may exist [30]. In this present study, the results of biological analysis shows that most of the proteins related to the cellular growth and proliferation functions as well as the invasion and proliferation abilities of tumor cells were down-regulated (Figure 4, Additional file 5: Figure s3, Additional file 6: File s5 and Additional file 7: File s6), demonstrating these functions may have been significantly inhibited. Except for HNRNPK and ELAVL1, another two candidate proteins, CDH2 and CALU, involved in the network of cellular growth and proliferation were selected and validated.
A previous study on brainstem glioma showed that higher expression of CDH2 predicts the progression of malignant tumors and tends to predict a shorter survival time of patients [31]. Other studies also indicated CDH2 may be functionally correlated with tumorigenesis in glioma cells and involved in mediating glioma cell migration [32-34]. In the present study, CDH2 is involved in several cellular functions (Additional file 5: Figure S3, Additional file 9: Table S3) and found to be down-regulated in PsPDs compared with GBMs (Table 1, Figure 4). The results were in accordance with previous studies and may demonstrate the malignancy changes in PsPD.
Another protein CALU, is a calcium-binding protein located in the endothelium that is involved in protein folding and sorting. This protein was recently found to be highly expressed in normal neural stem cells and GBM stem-like cells compared with the GBM tumor tissue [35]. Additionally, the gene CALU was also observed to be up-regulated in GBM but not in low-grade astrocytoma or oligodendroglioma [36]. These results indicated that the expression levels of CALU may be correlated with tumor cell proliferation ability, which is in accordance with the biological analysis results of this present study.
Validation of secretory proteins as candidate biomarkers
At present, there are no suitable specific biomarkers that can be used to accurately differentiate PsPDs from GBMs. Secretory proteins have the potential to be detected as biomarkers in body fluids. Therefore, we also selected three candidate secretory proteins, CALU (described above), FGB and FBLN1, for validation. The validation results were in accordance with the proteomic findings. It is noteworthy that, previous studies have reported that FBLN1 expression is elevated in breast tumors [37] and ovarian cancer cells [38]. But no details about the roles of FBLN1 in gliomas have been reported previously.
Taken together, the proteomic results as well as the validation results both identified that the expression level of HNRNPK, ELAVL1, CDH2 and FBLN1 in PsPDs were significantly different from GBMs (Figures 5, 6). ROC curves yielded an AUC value of 0.90 (95% CI, 0.662-0.9880) for CDH2 and.0.92 (95% CI, 0.696-0.995) for CDH2 combined with ELAVL1, which indicated that these two proteins could be potential biomarkers with relatively high accuracy in the diagnosis of PsPD.
Conclusion
In summary, our work offers an initial description of the proteins conserved in PsPDs and GBMs as well as novel information on proteins that are differentially expressed between groups. Through biological analysis and validation of the proteomic findings, this present study not only revealed the molecular signatures but also provide novel markers that may help to identify the mechanisms behind and allow the diagnosis of PsPD. However, due to the low number of samples used in the present study, above conclusions were just preliminary results, therefore, it should be careful to use our conclusions. Further verification in additional samples should be helpful and essential to understand the process.
Materials and methods
Sample collection and pathological examination
A set of fresh frozen tissue samples that included PsPD (n = 3) and newly diagnosed GBM (N-GBM, n = 3) was obtained under an Institutional Review Board-approved protocol at the Beijing Tiantan Hospital of Capital Medical University. Consents of clinical data and samples used for the study have been obtained from the patients and their families. PsPD was diagnosed according to the criteria of Macdonald [39] without viable tumor recurrence by pathological verification. The tissue samples were snap-frozen immediately after resection and stored at −80°C. To ensure that the fragments used for proteomic analysis contained a sufficient proportion (at least 80%) of the target tissue, we evaluated each specimen before use. Moreover, twelve additional samples were selected for verification by IHC and WB, including four PsPD, four N-GBM and four recurrent GBM (R-GBM) tissue samples (Additional file 9: Table S4).
ITRAQ sample preparation
First, 80 mg samples from each of the six frozen tissue samples selected for the proteomics screening were rinsed with PBS, and each sample was then mixed with lysis buffer (50 mMTris-HCl, 2.5 M thiourea, 8 M urea, 4% CHAPS, 65 mM DTT) for total protein extraction. The total protein concentration of each sample was determined using the Bio-Rad RC DC Protein Assay.
The proteins from each sample were pooled equally according to the total amount of protein and digested by filter-aided sample preparation combined with a microwave-assisted protein preparation method as previously described [40,41]. The peptides were dried by vacuum centrifugation and stored at −80°C.
The digested PsPD and GBM samples were mixed equally to create the internal standard and labeled by 114 iTRAQ. The three PsPD samples and the three GBM samples, were individually labeled with 115, 116 or 117 iTRAQ according to the manufacturer’s protocol (ABsciex).
2D-LC and MS/MS conditions
For offline separation a HPLC from Waters was used, and for online LC/MS/MS analysis a nano-ACQUITYUPLC system from Waters was used. First, the pooled mixture of the labeled samples was fractionated using a high-pH RPLC column from Waters (4.6 mm × 250 mm, C18, 3 μm). For each fraction the injection volume was 8uL. The samples were loaded onto the column in buffer A1 (1‰ aqueous ammonia in water, pH = 10), and eluted by buffer B1 (1‰ aqueous ammonia in 10% water and 90%ACN; pH = 10, flow rate = 1 mL/min) with the gradient of 5–90%for 60 min. The eluted peptides were collected at a rate of one fraction per minute, and pooled into 20 samples. Each sample was analyzed by LC-MS/MS using an RP C18 self-packing capillary LC column (75 μm × 100 mm, 3 μm) and a Triple TOF 5600 mass spectrometer. For Triple TOF 5600 a nano source was used. The MS data were acquired in high sensitivity mode with detailed parameters for Triple TOF 5600 being set as following: ion spray voltage was 2200v, curtain gas was 25, gas 1 was 5, gas 2 was 0, temperature was 150, declustering potential was 100, mass range was 350–1250 for MS and 100–1800 for MS/MS, collision energy was 35, and the resolution of MS and MS/MS was 40000 and 20000. An elution gradient of 5–30% buffer B2 (0.1% formic acid, 99.9% ACN; flow rate, 0.3 μL/min) for 50 min was used for the analysis. Thirty data-dependent MS/MS scans were acquired for every full scan. The normalized collision energy used was 35%, and charge state screening (including precursors with +2 to +4 charge state) and dynamic exclusion (exclusion duration of 15 s) were performed. Analyst TF 1.6 was used to control the instruments.
Database search
The MS/MS spectra were searched against the human subset of the Uniprot database (84910 entries) (http://www.uniprot.org/) using the Mascot software version 2.3.02 (Matrix Science, UK). Trypsin was chosen for cleavage with a maximum number of allowed missed cleavages of two. Carbamidomethylation (C) and iTRAQ 4-plex labels were set as fixed modifications. The searches were performed using a peptide and product ion tolerance of 0.05 Da. Scaffold software was used to further filter the database search results using the decoy database method with the following filter: a 1% false-positive rate at the protein level and two unique peptides per protein. After filtering the results as described above, the peptide abundances in different reporter ion channels of the MS/MS scan were normalized. The protein abundance ratio was based on unique peptide results. Proteins with a fold change ≥ 2 were considered significantly altered.
Bioinformatics analysis
Data filtering was performed according to strict criteria, wherein any missing data values or detection failures were deleted. Pearson’s correlation coefficient was calculated to measure the quantitative correlation among the three biological replicates in each group, and the coefficient of variation within groups was set at CV = 0.37 to filter out low-quality data. A Student’s t-test was performed between groups, and differences were considered to be significant when p < 0.05. Any proteins that satisfied the criteria of a fold change (FC) between groups of ≥2 were selected for bioinformatics analysis using Gene Ontology (GO) and Ingenuity Pathway Analysis (IPA).
GO functional and IPA network analysis
All proteins identified by the two approaches were assigned a gene symbol using the Panther database (http://www.pantherdb.org/). Protein classification was performed based on the functional annotations of the GO project for cellular compartment, molecular functional and biological processed. When more than one assignment was available, all of the functional annotations were considered in the results. Moreover, all of the selected proteins with a significant fold changes were used for pathway analysis using the IPA software (Ingenuity Systems, Mountain View, CA) for network analysis.
Immunohistochemistry and western blot analysis
IHC was performed on the same six tissue samples used for the proteomic analysis and on twelve additional formalin fixed, paraffin embedded tissue samples. The following primary antibodies were used: anti-ELAVL1mouse monoclonal (Santa Cruz), 1:500; anti-HNRNPK mouse monoclonal (Santa Cruz), 1:50;anti-CDH2rabbit monoclonal (Cell Signaling Technology), 1:250; anti-FBLN1 mouse monoclonal (Santa Cruz),1:125; anti-CALU goat polyclonal (Santa Cruz), 1:100; anti-FGB goat polyclonal (Abcam), 1:16000. After deparaffinization and rehydration, antigen retrieval was performed by immersing the slide in antigenretrieval buffer (10 mM sodium citrate, 0.05% Tween 20, pH = 6.0) at 95°C for 5 min using pressure cooker. Endogenous peroxidases were blocked with 0.03% hydrogen peroxide, and nonspecific binding was blocked with 2% fetal calf serum in Tris-buffered saline with 0.1% Triton X-100 (TBST, pH = 7.6). The sections were then incubated for 1 h at room temperature with primary antibodies followed by peroxidase-labeled polymer conjugate to anti-mouse, anti-rabbit, anti-goat immunoglobulins for 1 h and developed with diaminobenzidine system. The sections were counter stained with the Mayer’s hematoxylin and dehydrated, and the image was taken under microscope.
WBs of the same six samples and additional twelve samples was performed to validate the proteomic quantitation of four selected candidate proteins (HNRNPK, ELAVL1, CDH2 and FBLN1). Proteins extracted from GBM or PsPD tissues were resolved by SDS-PAGE (4–20% gradient precast gel; Invitrogen). The protein bands were electro transferred to a PVDF membrane (Millipore, Bedford, MA), blocked with 2% (v/v) BSA in TBST (150 mM NaCl, 20 mM Tris, 0.1% Tween 20, pH = 7.4) for 2 h at room temperature, followed by incubation with primary antibody (anti-ELAVL1, 1:200 (mouse monoclonal, Santa Cruz); anti-HNRNPK,1:3000 (mouse monoclonal, Santa Cruz); anti-CDH2, 1:800 (rabbit monoclonal, Cell Signaling Technology); anti-FBLN1, 1:100, (mouse monoclonal, Santa Cruz)) diluted with 1% BSA in TBST at room temperature for 2 h. After extensive wash with TBST, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody (anti-mouse or anti-rabbit; EarthOX, USA) diluted with 1% BSA in TBST for 90 min at room temperature. The membranes were developed using Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore). All the selected proteins ELAVL1, HNRNPK, CDH2 and FBLN1 were validated by Western blot analysis with actin as loading control.
Abbreviations
- 18 F-FDG:
-
18fluorodeoxyglucose
- CALU:
-
Calumenin
- CDH2:
-
N-cadherin
- CV:
-
Coefficient of Variance
- ELAVL1:
-
Hu-antigen R
- FBLN1:
-
Fibulin-1
- FC:
-
Fold Change
- FGB:
-
Fibrinogen Beta Chain
- GBM:
-
Glioblastoma
- GO:
-
Gene Ontology
- HNRNPK:
-
heterogeneous nuclear ribonucleoprotein K
- IDH1:
-
Isocitrate Dehydrogenase 1
- IHC:
-
Immunohistochemistry
- IPA:
-
Ingenuity Pathway Analysis
- MGMT:
-
O6-methylguanine–DNA methyltransferase
- MRI:
-
Magnetic Resonance Imaging
- MRS:
-
Magnetic Resonance Spectroscopy
- N-GBM:
-
Newly Diagnosed Glioblastoma
- Panther:
-
Protein Analysis Through Evolutionary Relationships
- PsPD:
-
Pseudoprogression Disease
- rCBV:
-
Relative Cerebral Blood Volume
- R-GBM:
-
Recurrent Glioblastoma
- VEGF:
-
Vascular Endothelial Growth Factor
- WB:
-
Western Blot
References
Hoffman WF, Levin VA, Wilson CB. Evaluation of malignant glioma patients during the postirradiation period. J Neurosurg. 1979;50:624–8.
De Wit M, De Bruin H, Eijkenboom W, Smitt PS, Van den Bent M. Immediate post-radiotherapy changes in malignant glioma can mimic tumor progression. Neurology. 2004;63:535–7.
Brandes AA, Tosoni A, Spagnolli F, Frezza G, Leonardi M, Calbucci F, et al. Disease progression or pseudoprogression after concomitant radiochemotherapy treatment: pitfalls in neurooncology. Neuro-Oncology. 2008;10:361–7.
Chaskis C, Neyns B, Michotte A, De Ridder M, Everaert H. Pseudoprogression after radiotherapy with concurrent temozolomide for high-grade glioma: clinical observations and working recommendations. Surg Neurol. 2009;72:423–8.
Topkan E, Topuk S, Oymak E, Parlak C, Pehlivan B. Pseudoprogression in patients with glioblastoma multiforme after concurrent radiotherapy and temozolomide. Am J Clin Oncol. 2012;35:284–9.
Chamberlain MC. Pseudoprogression in glioblastoma. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26:4359. author reply 4359–60.
Van Mieghem E, Wozniak A, Geussens Y, Menten J, De Vleeschouwer S, Van Calenbergh F, et al. Defining pseudoprogression in glioblastoma multiforme. European J Neurol Off J European Federation Neurol Soc. 2013;20:1335–41.
Kang HC, Kim CY, Han JH, Choe GY, Kim JH, Kim IA. Pseudoprogression in patients with malignant gliomas treated with concurrent temozolomide and radiotherapy: potential role of p53. J Neuro-Oncol. 2011;102:157–62.
Radbruch A, Fladt J, Kickingereder P, Wiestler B, Nowosielski M, Baumer P, et al. Pseudoprogression in patients with glioblastoma: clinical relevance despite low incidence. Neuro Oncol. 2015;17:151–9.
Gahramanov S, Muldoon LL, Varallyay CG, Li X, Kraemer DF, Fu R, et al. Pseudoprogression of glioblastoma after chemo- and radiation therapy: diagnosis by using dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging with ferumoxytol versus gadoteridol and correlation with survival. Radiology. 2013;266:842–52.
Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol Off J Am Soc Clin Oncol. 2010;28:1963–72.
Tsien C, Galbán CJ, Chenevert TL, Johnson TD, Hamstra DA, Sundgren PC, et al. Parametric response map as an imaging biomarker to distinguish progression from pseudoprogression in high-grade glioma. J Clin Oncol. 2010;28:2293–9.
Plotkin M, Eisenacher J, Bruhn H, Wurm R, Michel R, Stockhammer F, et al. 123I-IMT SPECT and 1HMR-spectroscopy at 3.0 T in the differential diagnosis of recurrent or residual gliomas: a comparative study. J Neuro-Oncol. 2004;70:49–58.
Van Laere K, Ceyssens S, Van Calenbergh F, de Groot T, Menten J, Flamen P, et al. Direct comparison of 18F-FDG and 11C-methionine PET in suspected recurrence of glioma: sensitivity, inter-observer variability and prognostic value. Eur J Nucl Med Mol Imaging. 2005;32:39–51.
Terakawa Y, Tsuyuguchi N, Iwai Y, Yamanaka K, Higashiyama S, Takami T, et al. Diagnostic accuracy of 11C-methionine PET for differentiation of recurrent brain tumors from radiation necrosis after radiotherapy. J Nucl Med. 2008;49:694–9.
Popperl G, Gotz C, Rachinger W, Gildehaus FJ, Tonn JC, Tatsch K. Value of O-(2-[18F] fluoroethyl)- L-tyrosine PET for the diagnosis of recurrent glioma. Eur J Nucl Med Mol Imaging. 2004;31:1464–70.
Brandes AA, Franceschi E, Tosoni A, Blatt V, Pession A, Tallini G, et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008;26:2192–7.
Motegi H, Kamoshima Y, Terasaka S, Kobayashi H, Yamaguchi S, Tanino M, et al. IDH1 mutation as a potential novel biomarker for distinguishing pseudoprogression from true progression in patients with glioblastoma treated with temozolomide and radiotherapy. Brain Tumor Pathol. 2013;30:67–72.
Pouleau HB, Sadeghi N, Baleriaux D, Melot C, De Witte O, Lefranc F. High levels of cellular proliferation predict pseudoprogression in glioblastoma patients. Int J Oncol. 2012;40:923–8.
Turtoi A, Musmeci D, Naccarato AG, Scatena C, Ortenzi V, Kiss R, et al. Sparc-like protein 1 is a new marker of human glioma progression. J Proteome Res. 2012;11:5011–21.
Mustafa DA, Dekker LJ, Stingl C, Kremer A, Stoop M, Smitt PAS, et al. A proteome comparison between physiological angiogenesis and angiogenesis in glioblastoma. Mol Cell Proteomics. 2012;11(M111):008466.
Lynch M, Chen L, Ravitz MJ, Mehtani S, Korenblat K, Pazin MJ, et al. hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor 4E (eIF4E) promoter, and its regulation of eIF4E contributes to neoplastic transformation. Mol Cell Biol. 2005;25:6436–53.
Carpenter B, MacKay C, Alnabulsi A, MacKay M, Telfer C, Melvin WT, et al. The roles of heterogeneous nuclear ribonucleoproteins in tumour development and progression. Biochimica et Biophysica Acta (BBA)-Reviews on. Cancer. 2006;1765:85–100.
Moumen A, Magill C, Dry KL, Jackson SP. ATM-dependent phosphorylation of heterogeneous nuclear ribonucleoprotein K promotes p53 transcriptional activation in response to DNA damage. Cell Cycle. 2013;12:698–704.
Strozynski J, Heim J, Bunbanjerdsuk S, Wiesmann N, Zografidou L, Becker SK, et al. Proteomic identification of the heterogeneous nuclear ribonucleoprotein K as irradiation responsive protein related to migration. J Proteomics. 2015;113:154–61.
Ido K, Nakagawa T, Sakuma T, Takeuchi H, Sato K, Kubota T. Expression of vascular endothelial growth factor-A and mRNA stability factor HuR in human astrocytic tumors. Neuropathol Off J Japanese Soc Neuropathol. 2008;28:604–11.
Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–39.
Fischer I, Gagner JP, Law M, Newcomb EW, Zagzag D. Angiogenesis in gliomas: biology and molecular pathophysiology. Brain Pathol. 2005;15:297–310.
Jensen RL. Brain tumor hypoxia: tumorigenesis, angiogenesis, imaging, pseudoprogression, and as a therapeutic target. J Neuro-Oncol. 2009;92:317–35.
da Cruz LC Jr H, Rodriguez I, Domingues RC, Gasparetto EL, Sorensen AG. Pseudoprogression and pseudoresponse: imaging challenges in the assessment of posttreatment glioma. AJNR Am J Neuroradiol. 2011;32:1978–85.
Wu W, Tian Y, Wan H, Ma J, Song Y, Wang Y, et al. Expression of beta-catenin and E- and N-cadherin in human brainstem gliomas and clinicopathological correlations. Int J Neurosci. 2013;123:318–23.
Kohutek ZA, Redpath GT, Hussaini IM. ADAM-10-mediated N-cadherin cleavage is protein kinase C-α dependent and promotes glioblastoma cell migration. J Neurosci. 2009;29:4605–15.
Péglion F, Etienne-Manneville S. N-cadherin expression level as a critical indicator of invasion in non-epithelial tumors. Cell Adhes Migr. 2012;6:327–32.
Barami K, Lewis-Tuffin L, Anastasiadis PZ. The role of cadherins and catenins in gliomagenesis. Neurosurg Focus. 2006;21:1–4.
Thirant C, Galan-Moya EM, Dubois LG, Pinte S, Chafey P, Broussard C, et al. Differential proteomic analysis of human glioblastoma and neural stem cells reveals HDGF as a novel angiogenic secreted factor. Stem Cells. 2012;30:845–53.
Sreekanthreddy P, Srinivasan H, Kumar DM, Nijaguna MB, Sridevi S, Vrinda M, et al. Identification of potential serum biomarkers of glioblastoma: serum osteopontin levels correlate with poor prognosis. Cancer Epidemiol Biomark Prev. 2010;19:1409–22.
Greene LM, Twal WO, Duffy MJ, McDermott EW, Hill AD, O’Higgins NJ, et al. Elevated expression and altered processing of fibulin-1 protein in human breast cancer. Br J Cancer. 2003;88:871–8.
Roger P, Pujol P, Lucas A, Baldet P, Rochefort H. Increased immunostaining of fibulin-1, an estrogen-regulated protein in the stroma of human ovarian epithelial tumors. Am J Pathol. 1998;153:1579–88.
Macdonald DR, Cascino TL, Schold Jr SC, Cairncross JG. Response criteria for phase II studies of supratentorial malignant glioma. J Clin Oncol Off J Am Soc Clin Oncol. 1990;8:1277–80.
Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62.
Sun W, Gao S, Wang L, Chen Y, Wu S, Wang X, et al. Microwave-assisted protein preparation and enzymatic digestion in proteomics. Mol Cell Proteomics. 2006;5:769–76.
Acknowledgements
This work was supported by the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2013BAI09B03) and Beijing Institute for Brain Disorders (BIBD-PXM2013_014226_07_000084).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declared that they have no competing interests.
Authors’ contributions
PZ carried out the sample preparation, proteomic analysis, biological analysis, sample validation using IHC and WB and manuscript drafting. ZG, DW, LZ participated in the 2D-LC analysis of samples and validations using WB. ZG, NJ, WS and LZ participate in the design of the study and the modification of the manuscript. WS and LZ both conceived of the study, and participated in the coordination. All authors read and approved the final manuscript.
Additional files
Additional file 1:
File s1. Quantitative Peptide List for PsPD Samples.
Additional file 2:
File s2. Quantitative Peptide List for N-GBM Samples.
Additional file 3:
File s3. Quantitative Protein List for PsPD Samples.
Additional file 4:
File s4. Quantitative Protein List for N-GBM Samples.
Additional file 5: Figure s1.
Pearson correlation coefficient plot of each two proteomic runs related to the tissue specimen in each group. The three graphs in the first row of the figure refers to Pearson coefficient of any two samples in PsPD sample group (ranged from 0.974 to 0.980); The three graphs in the second row of the figure refers to the Pearson coefficient of any two samples in GBM sample group (ranged from 0.967 to 0.978).
Additional file 6:
File s5. Significantly Fold Changed Proteins between PsPD and N-GBM Samples.
Additional file 7:
File s6. Quantitative peptides of differentially expressed proteins between PsPD and N-GBM samples.
Additional file 8:
File s7. List of secreted proteins.
Additional file 9: Table S1.
Parameters of ROC curve for four proteins.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Zhang, P., Guo, Z., Zhang, Y. et al. A preliminary quantitative proteomic analysis of glioblastoma pseudoprogression. Proteome Sci 13, 12 (2015). https://doi.org/10.1186/s12953-015-0066-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12953-015-0066-5