Abstract
Background
Artesunate plus sulfadoxine–pyrimethamine (ASP) is first-line treatment for uncomplicated Plasmodium falciparum malaria in most of India, except for six North-eastern provinces where treatment failure rates were high. In Ujjain, central India, the frequency of mutations associated with increased drug tolerance, but not overt resistance to sulfadoxine and pyrimethamine were 9% and > 80%, respectively, in 2009 and 2010, just prior to the introduction of ASP. The frequency of drug resistance associated mutations in Ujjain in 2015–2016 after 3–4 years of ASP use, are reported.
Methods
Blood samples from patients with P. falciparum mono-infection verified by microscopy were collected on filter-paper at all nine major pathology laboratories in Ujjain city. Codons pfdhfr 16–185, pfdhps 436–632 and K13 407–689 were identified by sequencing. Pfcrt K76T and pfmdr1 N86Y were identified by restriction fragment length polymorphism.
Results
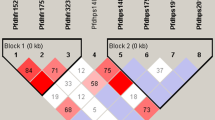
Sulfadoxine–pyrimethamine resistance-associated pfdhfr 108 N and 59R alleles were found in 100/104 (96%) and 87/91 (96%) samples, respectively. Pfdhps 437G was found in 10/105 (10%) samples. Double mutant pfdhfr 59R + 108 N were found in 75/81 (93%) samples. Triple mutant pfdhfr 59R + 108 N and pfdhps 437G were found in 6/78 (8%) samples. Chloroquine-resistance-associated pfcrt 76T was found in 102/102 (100%). Pfmdr1 N86 and 86Y were identified in 83/115 (72%) and 32/115 (28%) samples, respectively.
Conclusion
The frequency of P. falciparum with reduced susceptibility to sulfadoxine–pyrimethamine remained high, but did not appear to have increased significantly since the introduction of ASP. No polymorphisms in K13 associated with decreased artemisinin susceptibility were found. ASP probably remained effective, supporting continued ASP use.
Similar content being viewed by others
Background
Malaria caused an estimated 9.6 million infections and 16,700 deaths in India in 2017. Approximately 12% (163 million) and 81% (1.1 billion) of Indians live in high and low malaria transmission areas, respectively. Only 7% (88 million) live in areas considered to be malaria-free. Malaria thus remains a major cause of morbidity and mortality and threatens the majority of the population [1]. Moreover, malaria treatment is hampered by the spread and development of drug resistance in India as elsewhere in the world [2,3,4].
Chloroquine resistance arose in Southeast Asia and spread through India and Pakistan via the Northeastern states. Due to widespread chloroquine resistance, sulfalene-pyrimethamine followed by sulfadoxine–pyrimethamine (SP) monotherapy were officially recommended in Northeastern India from 1982. SP was subsequently replaced by artesunate plus sulfadoxine–pyrimethamine (ASP) in 2005, but this recommendation was discontinued in 2013 due to high treatment failure frequencies [3]. Artemether-lumefantrine (AL) is now recommended in several Northeastern states as well as in neighbouring Bangladesh. Due to spreading chloroquine resistance most of the remainder of India adopted ASP as first-line treatment by 2012. ASP is similarly recommended in neighbouring Pakistan [5]. There is thus considerable regional artemisinin and SP drug pressure and consequently a high risk of resistance spreading further. Continuous monitoring of ASP efficacy is therefore necessary [3].
Determining the frequency of genetic markers of SP resistance and reduced susceptibility to artemisnins is a rapid and easy way to monitor resistance. SP resistance is associated with mutations in the Plasmodium falciparum dihydrofolate reductase (pfdhfr) and dihyropteroate synthase (pfdhps) genes [6,7,8,9,10,11]. Accumulation of mutations in pfdhfr and pfdhps gradually increase the tolerability of P. falciparum to pyrimethamine and sulfadoxine, respectively [12]. Similarly, mutations in the Kelch 13 (K13) propeller domain of P. falciparum are associated with delayed parasite clearance during treatment with artemisnin derivatives. Specific genotypes in the chloroquine resistance transporter gene (pfcrt) and multidrug resistance gene (pfmdr 1) are associated with reduced susceptibility to lumefantrine [13,14,15,16,17].
The frequency of genotypes associated with SP and lumefantrine susceptibility in Ujjain, Madhya Pradesh, central India in 2009 and 2010, just prior to the introduction of ASP were reported previously [18]. The aim of this study was to determine the frequency of genotypes associated with reduced susceptibility to ASP and lumefantrine from the same area in samples collected 2015–2016, i.e. after 3–4 years of ASP use.
Methods
Study site and period
Ujjain district is located in the western part of Madhya Pradesh, central India. The population of the district is 1.9 million as per 2011 Census [19]. The climate is tropical and transmission may occur throughout the year provided the relative humidity levels support the vector survival. Ujjain district has low transmission of malaria with an annual parasite index (API) < 0.1 [20, 21]. Peak malaria transmission occurs during the warm and humid months of July to September. Data collection was from June to October in 2009, 2010, 2015 and 2016. Data from 2009 and 2010 were published previously and these samples used for K13 analyses only in the current study.
Recruitment of patients and sample collection
Samples and data were collected by the nine major pathology laboratories located in Ujjain city, Madhya Pradesh, India. Individuals or groups of pathologists reported the results from participating laboratories. Laboratories used microscopy to examine peripheral blood smears or rapid diagnostic tests (RDT) for the diagnosis of P. falciparum malaria. Inclusion criteria were microscopically or RDT verified malaria. For all patients that were smear positive for malaria, a drop of blood was put onto filter papers (Whatmann™ 3MM). RDTs were collected from patients in whom RDT only was used to diagnose malaria. RDTs or filter papers were labelled with the patient’s age and gender, dried and then placed inside individual sealed plastic bags.
Sample storage, DNA extraction
Filter papers and RDTs were stored at room temperature. DNA was extracted from two 3 mm Ø punches obtained from the filter paper or the whole RDT strips. DNA was extracted with Chelex®100 resin (Bio-Rad Laboratory, Hercules, CA) using the boiling method with minor modifications from the original protocol using 0.2% saponin/phosphate-buffered saline and 10% Chelex [22]. DNA was stored at − 20 °C until use.
Molecular analyses
Amino acid positions pfdhfr 16–185, pfdhps 436–632 and k13 407–689 were amplified using previously described PCR protocols and then sequenced commercially [13, 15, 23,24,25,26]. The Sequencher™ software version 4.6 (Gene Codes Corporation, Ann Arbor, MI, USA) was used for sequence analysis. The P. falciparum 3D7 clone sequences obtained from NCBI database were used as references for pfdhfr, pfdhps and k13.
Previously described multiplex PCR–RFLP (restriction fragment length polymorphism) methods with minor modifications were used to identify pfdhfr N51I, C59R and S108N and pfdhps S436F/A, A437G and K540E when sequencing failed and pfcrt K76T and pfmdr1 N86Y SNPs [23, 27, 28].
PCR and restriction products were resolved on 2% agarose gels (Amresco, Solon, OH, USA). All gels were stained with a nucleic acid gel stain (GelRed™, Biotium Inc, Hayward, CA, USA) and visualized under UV transillumination (GelDoc®, Biorad, Hercules, CA, USA). PCR products were purified and sequenced commercially (Macrogen Inc, Seoul, Korea).
Statistical analyses
The exact incidence of P. falciparum malaria was not known and we therefore decided to collect as many samples as possible over a two-year period. SNP frequencies were calculated by dividing the number of SNPs by the number of patients in whom a certain the allele could be identified. Allele frequencies in 2015 and 2016 were compared using Chi squared tests.
Ethics
Patients with uncomplicated malaria were enrolled and samples collected after informed oral consent of the patient or in the case of minors informed proxy-consent of their parent or guardian. The study was approved by the Institutional Ethics Committee of R D Gardi Medical College in Ujjain, Madhya Pradesh, India (61/2009 and 494/2016) and the Regional Ethics Committee in Stockholm, Sweden (2011/832-32/2).
Results
A total of 127 samples were collected during the 2015 (n = 48) and 2016 (n = 79) peak malaria transmission seasons. Plasmodium falciparum mono infection was identified in 124 samples and P. falciparum + Plasmodium vivax double infection were found in three samples. The samples were collected from 59 females and 68 males. The median age was 25 years, range 6 months to 76 years with no significant difference between females and males. Patients came from the following district Agar (n = 6), Badnagar (n = 2), Bhopal (n = 1), Dewas (n = 3, Indore (n = 2), Jhanua (n = 2), Mahidpur (n = 4), Rajgad (n = 1), Ratlam (n = 28), Shajapur (n = 8), Ahyamgad (n = 1), Tarana (n = 6) and Ujjain (n = 59). The district was not recorded for 3 patients. Blood for DNA extraction was available from filter-papers (n = 92) or rapid diagnostic tests (n = 27).
PCR success rate
The PCR success rate for each gene is shown in Table 1. PCR success was significantly (p < 0.001) lower if DNA was extracted from RDTs compared to filter-papers for all genes irrespective of whether RFLP or sequencing was used to identify SNPs. The difference was greatest for pfdhfr and pfdhps for which sequencing was only successful in 17–20% of RDT samples compared to 88 and 89% of filter-paper samples. Overall PCR success rate ranged from 69 to 91%.
Pfdhfr and Pfdhps SNP and haplotype frequencies
The number and frequencies of resistance-associated SNPs and haplotypes are shown in Tables 2 and 3, respectively. Double mutant pfdhfr 59R + 108 N were found in 78/87 (90%) samples. Triple mutant pfdhfr 50R + 59R + 108 N were found in 4/87 (5%) samples and double or triple were found in 82/87 (95%) samples.
Both pfdhfr and pfdhps haplotypes were successfully sequenced in 81 samples and triple mutant pfdhfr 59R + 108 N and pfdhps 437G were found in 8/83 (10%) samples. Quadruple mutant pfdhfr 50R + 59R + 108 N plus pfdhps 437G was found in 1/83 (1%) of samples. All these haplotypes also had the 436F allele, probably adding one resistance-associated SNP to each haplotype. No sample had resistance-associated alleles at pfdhfr codons 51 or 164 nor at pfdhps codons 540, 581 or 613.
The four samples with triple mutant pfdhfr 50R + 59R + 108 N were collected from four separate districts in 2015. Four of the ten pfdhps 437G (i.e. triple or quadruple mutant) mutant samples were collected in 2015 and six in 2016. Six of the ten samples came from Ujjain but all from different parts of Ujjain. There were no significant temporal or spatial trends.
Pfcrt K76T and pfmdr1 N86Y
Pfcrt 76T was detected in 100% (102/102) of samples. Pfmdr1 N86 was found in 72% (83/115) and pfmdr1 86Y in 28% (32/115) of samples. Pfmdr1 N86 was found in 10/10 samples with the pfdhps 437G allele and in 59/89 (66%) samples with the pfdhps 437A allele (p < 0.001).
K13 propeller region
The P. falciparum k13 propeller region was successfully sequenced in 92 samples collected in 2009 and 2010 prior to the introduction of ASP and in 87 samples collected in 2015 and 2016. Sensitive haplotypes only were found in all (n = 179) successfully sequenced samples.
Discussion
This is the second characterization of key anti-malarial drug resistance associated genetic polymorphisms in P. falciparum field isolates in Ujjain, Madhya Pradesh, Central India. Samples were collected 3 and 4 years after the implementation of ASP as first-line anti-malarial in Ujjain (2012) and 5 years after the first base-line study was conducted. From these data, the frequency of in vivo chloroquine and SP resistance in the study area and an indication of the speed at which resistant genotypes accumulate can be inferred.
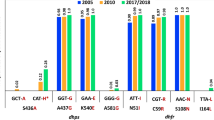
The most encouraging finding was that the frequency of the previously identified key SP resistance associated alleles had not increased significantly despite the presumed increased SP drug pressure. In the previous study, 90% (70/78) and 96% (75/78) of samples had pfdhfr 59R and 108 N mutations, respectively, compared to 96% in this study. Similarly, 100% (76/76) and 9% (7/77) of samples had the pfdhps 436F and 437G mutations, respectively in the previous study compared to 100% and 10% in the current study. Furthermore, the triple mutant pfdhfr 59R108N plus pfdhps 437G haplotype was also stable at a frequency of 10% in the current study compared to 8% (6/76) prior to introduction of ASP. As in the previous study no alleles at pfdhfr codons 51 or 164 nor at pfdhps codons 540, 581 or 613 that are associated with high levels of SP resistance were found. The results suggest that mutations associated with SP resistance have not accumulated rapidly since the introduction of ASP 3 to 4 years previously.
However, the majority of samples 68/83 had a double mutation (pfdhfr 59R + 108 N), triple mutations were found in 12% of samples and unlike the previous study a quadruple mutation (pfdhfr 50R59R108 N and pfdhps 437G) was found. Moreover, the triple pfdhfr 50R59R108 N, that was not seen prior to the introduction of ASP was seen in 5% of samples (4/87) possibly indicating that this haplotype is beginning to accumulate. In a more western part of Madhya Pradesh state, the pfdhfr 108 N, 59R and 51I frequencies were 80%, 57% and 32%, respectively and the frequency of triple pfdhfr mutations were 0%, 2%, and 3% in 2012, 2013 and 2014. The frequency of pfdhps double mutations were 0, 3 and 8.5% the same respective years [29]. The numbers are small but point in the same direction as our data possibly indicating that more resistant haplotypes are evolving in Madhya Pradesh. However, the risk of importing highly SP resistant P. falciparum is perhaps greater than the risk of local evolution as the frequency of pfdhfr + pfdhps with 6 or 7 mutations correlating to highly SP resistant parasites were 38% (85/226) in a study conducted in 2014–2016 only slightly further west, in West Bengal [2].
In the same West Bengal study various pfdhfr and pfdhps genotypes were tested for in vitro susceptibility [2]. Approximately half the parasites with pfdhfr 59R108N had IC50 values suggestive of pyrimethamine resistance. Samples with pfdhps 437G or 436A had sulfadoxine IC50 values ranging up to resistance level. Half the samples with 436A437G had IC50 values suggesting resistance. The effect of the pfdhps 436F mutation only on IC50 values was not assessed but has previously been shown to modulate sulfadoxine susceptibility [30]. The 82% frequency of pfdhfr 59R108N plus pfdhps 436F and the 11% frequency pfdhfr 59R108N plus pfdhps 436F437G thus suggests that the parasites are at least SP tolerant verging on resistant at our site. In line with these data, triple mutation pfdhfr 59R108N + pfdhps 437G have been associated with SP treatment failure in India [31].
Similar to findings in western Madhya Pradesh and Southwestern India but unlike findings in Northeastern India and West Bengal there were no mutations in the kelch-13 propeller domain suggesting that parasites remained artemisinin susceptible at the study site [2, 29, 32, 33]. ASP efficacy was 99.6% in the Madhya Pradesh study and 84% in the West Bengal study [2, 29]. The lack of mutations linked to delayed parasite clearance when treated with artemisinin or high degree of SP resistance suggests that the ASP efficacy at our site should be closer to the 99% found in Madhya Pradesh. The results thus support the continued use of ASP at the study site assuming continued direct or indirect monitoring of ASP efficacy.
In this study and in a previous report from the same study site pfdhps 436F was fixed [18]. Interestingly it was not noted further east in Madhya Pradesh but even further east in Orissa, the pfdhps F436A437K540A581A613 haplotype was found in 52% of samples [34]. Furthermore, the F436A437K540A581A613 and F436G437K540A581A613 haplotypes were found in 44% and 21% of samples at the same time as 33% carried the S436A437K540A581A613 haplotypes in study from Kolkata [35]. The 436F allele is thus not uncommon in India and its frequency varies.
The evolution of SP resistance in Asia has been shown to be ordered [12]. It appears to start with two initial mutations in pfdhfr (108N and then 59R), followed by two in pfdhps 437G and then either 540E or 581G. A third mutation then accumulates in each of pfdhfr and pfdhps [12]. In Ujjain, pfdhfr 50R appears to be developing in P. falciparum that already have pfdhfr 108N59R and pfdhps 436F concurrently with development of the pfdhfr (108N and then 59R) pfdhps 436F437G haplotype at our site. The numbers are small and no conclusions should be made. However, the pfdhps 436F allele has been shown to modulate sulfadoxine susceptibility [30] and was not detected in the study assessing the ordered evolution of SP resistance causing SNPs [12]. Its potential effect in the ordered accumulation of SNPs may thus be significant thought this has not been shown.
The 100% frequency of pfcrt 76T is significantly higher compared to the 96% (80/84) frequency prior to introduction of ASP (fishers exact p = 0.04). Similarly, though not significantly the pfmdr1 86Y frequency increased from 16% (13/83) prior to ASP introduction to 28% in the current study. This clearly indicates that chloroquine is not a viable treatment option. The pfcrt 76T and pfmdr1 86Y alleles are linked to lower lumefantrine IC 50 values suggesting that artemether–lumefantrine is a good second-line treatment in the case of ASP failure.
Finally, the poor PCR outcome in DNA extracted from RDTs is in line with other studies indicating the difficulty of extracting sufficient DNA [36]. Optimally blood for this type of survey should, therefore, be collected by other means so as not to introduce potential bias such as not being able to analyse samples with low parasite density.
Conclusions
No k13 SNPs were found and the frequency of SNPs associated with SP treatment failure was virtually unchanged 3–4 years after compared to before the introduction of ASP in Ujjain, Madhya Pradesh, Central India. However, a single quadruple mutation was found and a novel pfdhfr triple mutation was found in 5% of samples. The results support the continued use of ASP at the study site but indicate that continuous monitoring is necessary.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AL:
-
Artemether–lumefantrine
- ASP:
-
Artesunate plus sulfadoxine–pyrimethamine
- PCR:
-
Polymerase chain reaction
- Pfcrt P. falciparum :
-
Chloroquine resistance transporter
- Pfdhfr P. falciparum :
-
Dihydrofolate reductase
- Pfdhps P. falciparum :
-
Dihyropteroate synthase
- Pfmdr1 P. falciparum :
-
Multi drug resistance gene 1
- K13 :
-
Kelch 13
- RFLP:
-
Resitriction fragment length polymorphism
- SP:
-
Sulfadoxine–pyrimethamine
References
WHO. World Malaria Report Country Profiles. Geneva: World Health Organization; 2019.
Das S, Manna S, Saha B, Hati AK, Roy S. Novel pfkelch13 gene polymorphism associates with artemisinin resistance in Eastern India. Clin Infect Dis. 2019;69:1144–52.
Mishra N, Kaitholia K, Srivastava B, Shah NK, Narayan JP, Dev V, et al. Declining efficacy of artesunate plus sulphadoxine-pyrimethamine in northeastern India. Malar J. 2014;13:284.
Shah NK, Dhillon GP, Dash AP, Arora U, Meshnick SR, Valecha N. Antimalarial drug resistance of Plasmodium falciparum in India: changes over time and space. Lancet Infect Dis. 2011;11:57–64.
Yaqoob A, Khattak AA, Nadeem MF, Fatima H, Mbambo G, Ouattara A, et al. Prevalence of molecular markers of sulfadoxine–pyrimethamine and artemisinin resistance in Plasmodium falciparum from Pakistan. Malar J. 2018;17:471.
Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–13.
Foote SJ, Galatis D, Cowman AF. Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc Natl Acad Sci USA. 1990;87:3014–7.
Peterson DS, Milhous WK, Wellems TE. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci USA. 1990;87:3018–22.
Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–8.
Plowe CV, Kublin JG, Doumbo OKP. falciparum dihydrofolate reductase and dihydropteroate synthase mutations: epidemiology and role in clinical resistance to antifolates. Drug Res Updat. 1998;1:389–96.
Triglia T, Menting JG, Wilson C, Cowman AF. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997;94:13944–9.
Mita T, Ohashi J, Venkatesan M, Marma AS, Nakamura M, Plowe CV, et al. Ordered accumulation of mutations conferring resistance to sulfadoxine–pyrimethamine in the Plasmodium falciparum parasite. J Infect Dis. 2014;209:130–9.
Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–5.
Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–71.
Malmberg M, Ferreira PE, Tarning J, Ursing J, Ngasala B, Bjorkman A, et al. Plasmodium falciparum drug resistance phenotype as assessed by patient anti-malarial drug levels and its association with pfmdr1 polymorphisms. J Infect Dis. 2013;207:842–7.
Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, et al. In vivo selection of Plasmodium falciparum pfmdr1 86 N coding alleles by artemether-lumefantrine (Coartem). J Infect Dis. 2005;191:1014–7.
Mwai L, Kiara SM, Abdirahman A, Pole L, Rippert A, Diriye A, et al. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob Agents Chemother. 2009;53:5069–73.
Pathak A, Mårtensson A, Gawariker S, Mandliya J, Sharma A, Diwan V, et al. Characterization of drug resistance associated genetic polymorphisms among Plasmodium falciparum field isolates in Ujjain, Madhya Pradesh, India. Malar J. 2014;13:182.
Ujjain District: Census 2011 data http://www.census2011.co.in/census/district/302-ujjain.html.
Annual Parasite Index, Madhya Pradesh http://nvbdcp.gov.in/images/MadhyaPrd.jpg.
Programme NVBDC. Malaria Situation. 2019.
Wooden J, Kyes S, Sibley CH. PCR and strain identification in Plasmodium falciparum. Parasitol Today. 1993;9:303–5.
Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–63.
Vinayak S, Alam MT, Mixson-Hayden T, McCollum AM, Sem R, Shah NK, et al. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathol. 2010;6:e1000830.
Alam MT, de Souza DK, Vinayak S, Griffing SM, Poe AC, Duah NO, et al. Selective sweeps and genetic lineages of Plasmodium falciparum drug-resistant alleles in Ghana. J Infect Dis. 2011;203:220–7.
Kone A, Mu J, Maiga H, Beavogui AH, Yattara O, Sagara I, Tekete MM, et al. Quinine treatment selects the pfnhe-1 ms4760-1 polymorphism in Malian patients with Falciparum malaria. J Infect Dis. 2013;207:520–7.
Veiga MI, Ferreira PE, Bjorkman A, Gil JP. Multiplex PCR-RFLP methods for pfcrt, pfmdr1 and pfdhfr mutations in Plasmodium falciparum. Mol Cell Probes. 2006;20:100–4.
Ursing J, Kofoed PE, Rodrigues A, Rombo L, Gil JP. Plasmodium falciparum genotypes associated with chloroquine and amodiaquine resistance in Guinea-Bissau. Am J Trop Med Hyg. 2007;76:844–8.
Mishra S, Bharti PK, Shukla MM, Ali NA, Kashyotia SS, Kumar A, et al. Clinical and molecular monitoring of Plasmodium falciparum resistance to antimalarial drug (artesunate + sulphadoxine − pyrimethamine) in two highly malarious district of Madhya Pradesh, Central India from 2012–2014. Pathog Glob Health. 2017;111:186–94.
Ahmed A, Bararia D, Vinayak S, Yameen M, Biswas S, Dev V, et al. Plasmodium falciparum isolates in India exhibit a progressive increase in mutations associated with sulfadoxine–pyrimethamine resistance. Antimicrob Agents Chemother. 2004;48:879–89.
Mohapatra PK, Sarma DK, Prakash A, Bora K, Ahmed MA, Sarma B, et al. Molecular evidence of increased resistance to anti-folate drugs in Plasmodium falciparum in North-East India: a signal for potential failure of artemisinin plus sulphadoxine–pyrimethamine combination therapy. PLoS One. 2014;9:e105562.
Mishra N, Bharti RS, Mallick P, Singh OP, Srivastava B, Rana R, et al. Emerging polymorphisms in falciparum kelch 13 gene in Northeastern region of India. Malar J. 2016;15:583.
Wedam J, Tacoli C, Gai PP, Siegert K, Kulkarni SS, Rasalkar R, et al. Molecular evidence for Plasmodium falciparum resistance to sulfadoxine–pyrimethamine but absence of K13 mutations in Mangaluru, Southwestern India. Am J Trop Med Hyg. 2018;99:1508–10.
Ahmed A, Lumb V, Das MK, Dev V, Wajihullah, Sharma YD. Prevalence of mutations associated with higher levels of sulfadoxine–pyrimethamine resistance in Plasmodium falciparum isolates from Car Nicobar Island and Assam, India. Antimicrob Agents Chemother. 2006;50:3934–8.
Chatterjee M, Ganguly S, Saha P, Guha SK, Maji AK. Polymorphisms in pfdhfr and pfdhps genes after five years of artemisinin combination therapy (ACT) implementation from urban Kolkata, India. Infect Genet Evol. 2017;53:155–9.
Nag S, Ursing J, Rodrigues A, Crespo M, Krogsgaard C, Lund O, et al. Proof of concept: used malaria rapid diagnostic tests applied for parallel sequencing for surveillance of molecular markers of anti-malarial resistance in Bissau, Guinea-Bissau during 2014–2017. Malar J. 2019;18:252.
Acknowledgements
The authors acknowledge the help of Dr. GK Nagar, Dr. Girish Jarare, and Dr. Kishore Jhamnani in data collection. We thank all the patients for giving us consent to collect the blood samples.
Funding
No external funding was received. JU holds a clinical researcher position funded by Stockholm county council (Award number 20160597)
Author information
Authors and Affiliations
Contributions
AP, AM and JU conceived the study. AP organised data collection. AP, AM, SBG, AS, VD, MP, JU designed the methodology. AP, SBG, AS, VD and MP collected the field samples. JU conducted the molecular analyses. AP, JU and AM drafted the manuscript. SBG, AS, VD and MP provided valuable insights during the revision and editing of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Patients with uncomplicated malaria were enrolled and samples collected after informed oral consent of the patient or in the case of minors informed proxy-consent of their parent or guardian. The study was approved by the Institutional Ethics Committee of R D Gardi Medical College in Ujjain, Madhya Pradesh, India (61/2009 and 494/2016) and the Regional Ethics Committee in Stockholm, Sweden (2011/832-32/2).
Consent for publication
Not applicable.
Competing interests
All authors: No reported competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Pathak, A., Mårtensson, A., Gawariker, S. et al. Stable high frequencies of sulfadoxine–pyrimethamine resistance associated mutations and absence of K13 mutations in Plasmodium falciparum 3 and 4 years after the introduction of artesunate plus sulfadoxine–pyrimethamine in Ujjain, Madhya Pradesh, India. Malar J 19, 290 (2020). https://doi.org/10.1186/s12936-020-03274-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-020-03274-w