Abstract
Background
Bats are home to diverse haemosporidian parasites namely Plasmodium and Plasmodium-related. While information is available at a worldwide level, haemosporidian infection in bats from Madagascar is still scarce and recent changes in the taxonomy of the island’s bat fauna, particularly the description of several new species, require a reassessment of previously described patterns, including blood parasite ecology and vectorial transmission.
Methods
A sample representing seven of the nine known bat families and 31 of the 46 currently recognized taxa from Madagascar and collected in the western and central portions of the island were screened by PCR for the presence of Polychromophilus. In addition, Nycteribiidae flies parasitizing Miniopteridae and Vespertilionidae were screened for parasites with the aim to better understand aspects of vector transmission. Phylogenetic reconstruction using the mitochondrial cytochrome b encoding gene was used in a Bayesian analysis to examine the relationship between Polychromophilus recovered from Malagasy bats and those identified elsewhere.
Results
Polychromophilus infection was restricted to Miniopterus spp. (Miniopteridae), Myotis goudoti (Vespertilionidae), and Paratriaenops furculus (Rhinonycteridae), with an overall infection rate of 13.5%. Polychromophilus melanipherus was found infecting Miniopterus spp. and P. furculus, whereas Polychromophilus murinus was only recovered from M. goudoti. These two protozoan parasites species were also detected in bat flies species known to parasitize Miniopterus spp. and M. goudoti, respectively. Generalized linear model analyses were conducted to elucidate the effect of species and sex on haemoparasites infection in Miniopterus spp., which revealed that males have higher risk of infection than females and prevalence differed according to the considered Miniopterus host. Molecular screening of nycteribiid flies revealed three positive species for Polychromophilus spp., including Penicillidia sp. (cf. fulvida), Penicillidia leptothrinax, and Nycteribia stylidiopsis. These three fly species are known to parasitize Miniopterus spp. and M. goudoti and should be considered as potential vectors of Polychromophilus spp.
Conclusion
Phylogenetic analyses demonstrated the existence of at least four distinct clades within the genus Polychromophilus, two of which were documented in the present study. The screening of nycteribiid flies overlaid on the highly diversified genus Miniopterus, provides considerable insight into parasite transmission, with bat infection being associated with their roosting behaviour and the occurrence of specific arthropod vectors.
Similar content being viewed by others
Background
A number of different studies have been carried out to understand the evolutionary biology of haemosporidian parasites [1, 2]. The group Plasmodium, which infects humans, has been examined in detail because of its public health consequences [3, 4]. The genus Plasmodium has also been documented in different vertebrate groups, such as reptiles, birds and mammals, including non-human primates, based on morphological and molecular studies [1, 5, 6]. In other groups of vertebrates, malaria-related parasites are also known and these parasites have a common characteristic in their life cycle in that arthropods act as vectors. Malaria and malaria-related parasites form a paraphyletic groups within the haemosporidian and understanding the existing diversity is a key to having greater insight into their evolutionary biology.
As far as bats are concerned, their longevity, gregarious behaviour, dispersal potential and possible permissive immunity system have been proposed to facilitate parasite infection and maintenance [7]. Bats are hosts to different malaria and malaria-related parasites [6, 8,9,10,11]. To date, seven bat families are known to be infected by haemosporidian parasites: Hipposideridae, Megadermatidae, Miniopteridae, Pteropodidae, Rhinolophidae, Rhinonycteridae and Vespertilionidae [6, 8, 10, 12]. Different morphological characters associated with the taxonomy of bat malarial parasites have been previously published [13,14,15,16], and their systematics as well as species diversity have been clarified through molecular approaches [6, 9, 17, 18]. Thus far, based on morphological and molecular studies, eight genera of haemosporidian parasites are recognized to infect bats: Biguetiella, Dionisia, Hepatocystis, Johnsprentia, Nycteria, Plasmodium, Polychromophilus and Sprattiella [6, 8, 10, 15, 16, 18, 19]. With regards to the genus Polychromophilus, two species, Polychromophilus melanipherus and Polychromophilus murinus, were originally described from Miniopterus schreibersii and Vespertilio murinus, respectively [20]. These two haemosporidian taxa are known to infect bats belonging to the Miniopteridae and Vespertilionidae in Africa and Europe [9, 17]. Further, previous morphological descriptions identified three additional species, namely Polychromophilus deanei, Polychromophilus adami and Polychromophilus corradetti, infecting Myotis nigricans [13], Miniopterus minor and M. schreibersii, respectively [12]. In the literature, parasites infecting Myotis spp., Eptesicus fuscus and V. murinus have been placed within the genus Bioccala, formerly recognized as a subgenus of Polychromophilus [16, 21]; however, this has yet to be verified based on molecular studies.
As far as bats from Madagascar are concerned, considerable progress has been made in recent years regarding their systematics and taxonomy, and the fauna is currently composed of 46 recognized species of which nearly 80% are endemic [22,23,24,25,26]. Aspects of their roosting ecology and distribution have also been studied [23, 27]. Research examining blood parasite diversity in Malagasy bats using blood smear screening focused on 14 different bat species [28] and found the presence of Haemoproteidae in Miniopterus gleni and Miniopterus manavi sensu lato (Miniopteridae), Myotis goudoti (Vespertilionidae) and Paratriaenops furculus (formerly placed in the genus Triaenops) (Rhinonycteridae). Further, Duval et al. [8], reported the presence of Haemosporidia in M. manavi s.l. and M. goudoti without details on the taxonomy of these parasites. However, recent morphological and molecular studies revealed that these hemosporidian parasites are from two distinct clusters within Polychromophilus: P. melanipherus and P. murinus, detected in M. manavi s.l. and M. goudoti, respectively [17]. It is important to note that the number of recognized species within the genus Miniopterus on Madagascar has increased from four [22] to 12 [23, 24, 26] and M. manavi cited by Peterson and colleagues [22] represents a complex of paraphyletic species composed of at least eight cryptic taxa [23, 24, 26]. Further, different sister species relationships have been described within Malagasy members of this genus [29].
These different scientific advancements underline that available information on haemosporidian parasites infecting bats from Madagascar is scarce. Further, details on infection rates and host identity require further investigation to understand parasite ecology and distribution across the island.
Methods
Sampling sites and techniques
Fifty-two sites were visited from February 2012 to March 2013 in different areas of Madagascar to sample bats associated with taxonomic studies [25], ecology and distribution [27], as well as hosted ectoparasite and microparasites [30,31,32,33]. Bats were captured using mist nets and harp traps, which were most often placed at cave entrances or across bat flight pathways (Fig. 1). In a few cases, a butterfly net was used to sample individuals from cave and synanthropic day-roost sites. The exception was for Pteropus rufus, a CITES Appendix II species, for which living individuals were purchased in a market and not physically captured by the field research group. This species is considered as bushmeat on Madagascar and exportation of tissue samples for scientific work needed a CITES permits (cf. Ethical approval and consent to participate).
Upon capture, each individual bat, excluding Pteropus rufus as noted above, was placed in a clean cloth bag. Further, Nycteribiidae flies, an obligate parasite of bats [34] and parasitizing members of the families Miniopteridae and Vespertilionidae, were directly collected from individual bats for morphological and molecular identification [30, 33]. Voucher specimens were deposited at the Université d’Antananarivo, Mention Zoologie et Biodiversité Animale (formerly Département de Biologie Animale) (UADBA), Antananarivo, and at the Field Museum of Natural History (FMNH), Chicago.
DNA extraction and PCR screening
A pool of approximately 1 mm3 of kidney, lung and spleen from each individual bat was crushed in DMEM medium using two 3 mm-tungsten beads in a Tissue Lyser II (Qiagen, Valencia, CA). Subsequently, the mixture was centrifuged and the supernatant used for nucleic acids extraction. Total nucleic acids were extracted using EZ1 robot, with the viral mini kit v2.0 according to the manufacturer’s protocol (Qiagen Valencia, CA, USA), which has been routinely used in the PIMIT laboratory for the screening of both RNA and DNA parasites as previously described [32, 35, 36]. Nycteribiidae recovered from bats belonging to the families Miniopteridae and Vespertilionidae were crushed in a Tissue LyserII using two 3 mm tungsten beads, and total nucleic acids were extracted with the Qiagen biorobot with the viral mini kit v2.0 following manufacturer’s protocol (Qiagen Valencia, CA, USA) (see [30] for additional information).
Bat samples and Nycteribiidae flies were screened for the presence of Polychromophilus spp. using a previously described nested PCR protocol targeting the mitochondrial cytochrome b locus (cyt b) of haemosporidian parasites, which has been used to screen bats from Madagascar, Cambodia and Switzerland [8, 17, 18]. Primary PCR were conducted in a 25 µl reaction volume containing 12.5 µl of GoTaq® Hot start green master mix (Promega, Madison, WI, USA), 1 µl of each Plas1/Plas2 primer at 0.4 µM and 1 µl of total nucleic acid as template. The balance of the reaction volume was supplemented by nuclease free water. The cycling conditions were 40 s at 94 °C, 40 s at 50 °C, 1 mn at 72 °C for 35 cycles. Secondary PCRs were performed using 50 µl reactions containing 25 µl of GoTaq® Hot start, 1 µl of each Plas3/Plas4 primers, and 1 µl of the primary PCR product completed with nuclease free water. The cycling conditions were 40 s at 94 °C, 40 s at 50 °C, 1 mn at 72 °C, and a final extension at 72 °C for 7 mn. All PCR reactions were preceded by an initial denaturation at 94 °C for 5 mn. PCR products were visualized in an electrophoresis gel and sent to Genoscreen (Lille, France) for direct Sanger sequencing on both strands using forward and reverse primers.
Phylogenetic analyses
Sequences from positive individual of bats and nycteribiid flies were visually edited in Geneious 6.1.4 (http://www.geneious.com/) [37]. For positive individuals, the consensus nucleotide sequences from each were saved and used in the phylogenetic study. For similar consensus nucleotide sequences obtained from bats, the different haplotype sequences were identified using “pegas” package [38] implemented in R software [39] and only one sequence per haplotype was used in the analysis (Additional file 1: Table S1). Selected nucleotide sequences were then combined with those downloaded from GenBank and subsequently aligned using MAFFT implemented in Geneious software [37].
Prior to conducting phylogenetic analyses, a jModelTest version 2.1.3 [40, 41] was performed revealing GTR + G as the best substitution model. Subsequently, Bayesian inference consisting of two independent runs of four incremental Metropolis Coupled Markov Chain Monte Carlo (MC3) iterations starting from a random tree was conducted using MrBayes 3.1.2 [42]. This analysis was run for 5,000,000 generations with trees and associated model parameters sampled every 500 generations. The first 10% of the trees were discarded as a burn-in. New nucleotide sequences produced in the context of this study were deposited in GenBank under accession numbers MH744503 to MH744537 (Additional file 1: Table S1).
Morphological examination of blood parasites
In the field, a thin blood smear was prepared from each captured bat, specifically to examine haemoparasite morphology. Blood smears were fixed with methanol and stained with GIEMSA at room temperature. In the context of this study, blood smears were not used for detecting infection, as PCR analysis is distinctly more sensitive than visual inspection [17, 43,44,45]. Further, in many cases, morphology of apicomplexan parasites does not provide sufficient characters for species-level identification. Nevertheless, some blood smears of PCR positive samples were examined to document the morphology of associated haemosporidian parasites for illustrative purposes. In such cases, blood smears were examined using a binocular microscope under immersion oil objective with 1000× magnification.
Statistical analyses
Generalized Linear Model (GLM) analyses were conducted to investigate variation in Polychromophilus infection rate between species and sex classes in Miniopterus spp. and a Chi square analysis was conducted to compare infection rates between sex classes in M. goudoti. All statistical computations were conducted using R version 3.0.0 [39].
Results
In total, 947 individual bats belonging to 31 of the 46 species currently recognized taxa on Madagascar were tested for the presence of Polychromophilus spp. Three of the seven tested families—Miniopteridae, Vespertilionidae and Rhinonycteridae—were found positive based on molecular screening with a total infection rate of 13.5% (Table 1). Within the Miniopteridae, all tested species were positive with significant difference in infection rates between species, ranging from 25.8% in Miniopterus mahafaliensis to 72.7% in M. gleni. Further, males had a greater chance of being infected than females (Table 2). Within the Vespertilionidae, only M. goudoti was found positive and had an infection rate of 41.7%; the difference in infection rates between males and females in M. goudoti was not statistically significant (Pearson Chi square: X2 = 1.62, d.f. = 1, P = 0.203). Finally, within the Rhinonycteridae, only a single individual of P. furculus (7.1%) tested positive for Polychromophilus.
Phylogenetic analyses and host-parasite relationship
Phylogenetic analysis based on cyt b revealed that Polychromophilus infecting bats in different portion of the world forms a monophyletic clade composed of four clusters. Polychromophilus in Malagasy bats is segregated into two clusters. A first cluster is composed of P. melanipherus (Fig. 2a) lineages infecting all eight Miniopterus spp. from Madagascar screened in the present investigation, as well as a single individual of P. furculus (H1 to H24 in the phylogenetic tree, Fig. 3, Additional file 1: Table S1). Polychromophilus melanipherus identified from Malagasy Miniopteridae clustered with those identified in M. schreibersii from Switzerland, Miniopterus inflatus from Gabon, and Miniopterus villiersi from Guinea (Fig. 3). Hence, excluding P. furculus, P. melanipherus is known from the Miniopteridae of Africa, Europe, and Madagascar. The second cluster is composed of P. murinus (Fig. 2b) recovered from M. goudoti on Madagascar (H1 to H5 in the phylogenetic tree, Fig. 3, Additional file 1: Table S1). Although the Malagasy clade of P. murinus forms a monophyletic group with haemoparasites recovered from Myotis daubentonii and other European Vespertilionidae (Fig. 3), they are not embedded within the clade and show a certain level of genetic divergence. Haemosporidian parasites identified from Kerivoula hardwickii (Vespertilionidae) sampled in Cambodia and from Neoromicia capensis and Pipistrellus aff. grandidieri (Vespertilionidae) sampled in Guinea cluster in two distinct clades labelled Polychromophilus sp. 1 and 2, respectively (Fig. 3).
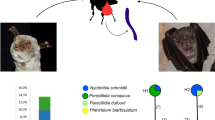
Bayesian reconstruction showing Polychromophilus spp. infecting Malagasy bats (in blue) and Nycteribiidae (in red) based on cytochrome b. Only values in the major nodes were represented for higher posterior probabilities (> 0.9). Polychromophilus melanipherus identified in Paratriaenops furculus are followed by an asterisk. Mad: Madagascar, Gui: Guinea, Sw: Switzerland, Gab: Gabon, Ple: Penicillidia leptothrinax, Psp: Penicillidia sp. (cf. fulvida), Nsty: Nycteribia stylidiopsis
Molecular screening of Nycteribiidae: insights on their role as potential vectors
In total, 38 individual nycteribiids belonging to three species (17 Penicillidia leptothrinax, 2 Penicillidia sp. (cf. fulvida), and 19 Nycteribia stylidiopsis) were tested via PCR screening for the presence of Polychromophilus. Five bat fly specimens were positive for P. melanipherus and one for P. murinus. Bat flies positive for P. melanipherus via the PCR analyses included four P. leptothrinax (two sampled on Miniopterus aelleni and two on M. manavi s.l.) in addition to a N. stylidiopsis obtained on M. gleni. Of note, this latter nycteribiid was sampled on a PCR negative bat host. Further, one Penicillidia sp. (cf. fulvida) specimen collected on Miniopterus griveaudi was found positive for P. murinus, whereas the associated bat host was infected with P. melanipherus.
Discussion
Malarial parasites are known to infect a wide diversity of vertebrates, specifically birds [43, 45, 46], reptiles [1, 47], and mammals [1], including bats [6, 17, 19]. This paraphyletic apicomplexan group is highly diverse with different speciation and host-switching events [1, 8]. Haemosporidian parasites infecting bats were first reported by Dionisi in 1899 [20] and an updated checklist of chiropteran haemosporidia has been recently published based on their gametocyte types and schizont locations [16]. With respect to Malagasy bats, a few studies have been conducted to detect and identify haemosporidian parasites in these animals based on morphological and molecular tools [8, 28]. However, a number of bat taxa have hitherto not been screened; a necessary step to understand infection patterns and identify biotic and abiotic parameters influencing parasite transmission.
Phylogenetic relationship of haemosporidian parasites
Previous phylogenetic studies of haemosporidian parasites infecting Malagasy bats revealed the presence of two different parasite species belonging to the genus Polychromophilus [6, 8, 17]. In this study, the presence of two Polychromophilus sister species infecting two widely distributed bat genera on Madagascar, namely Miniopterus (Miniopteridae) and Myotis (Vespertilionidae) was confirmed. Polychromophilus melanipherus infects all eight Miniopterus spp. sampled in the present study, which are endemic to the Malagasy Region (Madagascar and the Comoros). The haplotypes of haemoparasites infecting Malagasy bats are embedded within the P. melanipherus clade occurring in African and European Miniopterus spp. This topology strongly supports a tight specificity of P. melanipherus within hosts belonging to the Miniopteridae [6, 17, 48]. One individual of P. furculus (Rhinonycteridae) also tested positive and based on the cyt b sequence, it fell within the P. melanipherus clade. Raharimanga and colleagues [28] and Duval and colleagues [8], already reported the presence of one individual of P. furculus with haemosporidian infection based on microscopic examination. This P. furculus specimen might have been accidentally and transiently infected by an infected nycteribiid during a blood meal. The haplotype identified in this P. furculus is unique and different from haplotypes obtained from infected Miniopterus sampled in the same day roost. However, further investigations are needed to elucidate the importance of P. melanipherus infection in P. furculus with a greater sample from different localities.
The second cluster of Polychromophilus only infected M. goudoti and is genetically related to P. murinus. Interestingly, P. murinus from M. goudoti, a bat endemic to Madagascar, was not embedded within the cluster of P. murinus from European bat species and suggesting a different diversification history of these two subgroups. It has been suggested that the lineage in which M. goudoti is placed (Ethiopian clade V) diverged from the other lineages mainly composed of Palearctic and Oriental Myotis spp. (Clade I, II, III, IV) in the Miocene, over 11 Mya ago [49]. It can thus be hypothesized that P. murinus parasites infecting M. goudoti co-diverged with their host and infects a higher diversity of bats within the family Vespertilionidae such as Myotis daubentonii, Eptesicus serotinus, and Nyctalus noctula [8, 18].
No co-infection was found in the present study, which confirms previously published information [6, 17]. This suggests that infection is actually specific at the level of host genus. This pattern is remarkable in that on Madagascar Miniopterus spp. and M. goudoti are syntopic in the same day-roost sites. Such physical contact has been previously reported to favour host switching of pathogenic Leptospira between these two syntopic occurring host genera [32], but this does not seem to be the case for Polychromophilus spp.
Polychromophilus infection in bats from Madagascar
The molecular screening of Malagasy bats revealed a total infection rate of 13.5%, which is congruent with previously reported rates [8, 28]. Based on the analysis of 31 Malagasy bat taxa, haemosporidian infection is limited to the families Miniopteridae, Vespertilionidae, and Rhinonycteridae. Bayesian reconstruction showed the presence of P. melanipherus infecting all eight Miniopterus spp. sampled herein, as well as a single P. furculus specimen, while P. murinus was only detected in M. goudoti. While a significant difference was observed between sexes in P. melanipherus infecting Miniopterus spp., no such pattern was observed in P. murinus infecting M. goudoti. These results can be explained by the roosting ecology and behaviour of these two syntopic species. Both sexes of M. goudoti live together in colonies throughout the year. For Miniopterus spp., there is at least partial sexual segregation in day-roosting sites. For example, M. manavi s.l. was sampled on several occasions in an open rock-shelter cave at Ambohitantely, where only males were present, suggesting that the two sexes are in contact only during the initial stages of reproduction. Further, bats roosting in this shallow cave are presumably more exposed to haematophagous insects than bats of either sex roosting deep in caves. Nevertheless, for species such as M. mahafaliensis sampled in the present study and at different localities, both sexes were present within their day roost site.
Insight into invertebrate vectors
Polychromophilus spp. have been reported to be transmitted by nycteribiid flies [13, 14, 50], wingless Diptera that are obligate blood-sucking parasites of bats [34]. Molecular screening of bat flies sampled in the context of this study was carried out to identify potential candidate vectors. The screening results should be interpreted with some caution, as the presence of Polychromophilus DNA in a nycteribiid might simply be the result from a recent blood meal. Flies sample included P. leptothrinax, specific to Miniopterus spp., and Penicillidia sp. (cf. fulvida) and N. stylidiopsis, parasitizing both Myotis and Miniopterus [30, 33]. In several cases, Penicillidia sp. (cf. fulvida) and N. stylidiopsis were found on the same bat host [30, 33]. Both P. leptothrinax and N. stylidiopsis tested positive for P. melanipherus. Further, P. murinus was detected in a Penicillidia sp. (cf. fulvida) fly that was collected on a Miniopterus griveaudi specimen testing positive for P. melanipherus. Although, cross contamination cannot be excluded during bat sampling as Miniopterus griveaudi and M. goudoti occur in syntopic cave day-roost sites, the result can alternatively be interpreted as additional evidence for Miniopterus spp. being non-permissive to P. murinus infection. This situation may be related to the ecology of Miniopterus and Myotis. In fact, colonies of Miniopterus spp. and M. goudoti often live in syntopy (physical contact). Members of these two genera seem to have ectoparasites with relaxed host preference [30, 33]. However, given their close physical contact, it might be expected to favor parasite co-infection and/or host switching. However, no P. murinus/P. melanipherus co-infection was detected either in bat genera or in their nycteribiid ectoparasites [6, 17, 51], and thus suggesting host-specificity.
Conclusions
This work provides further advances regarding previous studies on the taxonomy and distribution of Polychromophilus spp. in bats occurring on Madagascar. While Polychromophilus infections seem to be mostly limited to the families Miniopteridae and Vespertilionidae, the presence of other apicomplexan parasites, such as Plasmodium and Hepatocystis, should also be investigated using molecular and morphological techniques. As a vector-borne infection, future work on haemosporidian parasites should carefully address the biology, ecology, and distribution of invertebrate vectors. The important diversity of Malagasy bats, especially the 12 currently recognized species of Miniopterus with different distributions and reproduction behaviour, together with the specificity of Polychromophilus make this biological model particularly suitable to investigate the impact of biotic and abiotic factors on the transmission of haemoparasites.
References
Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol Phylogenet Evol. 2008;47:261–73.
Loy DE, Liu W, Li Y, Learn GH, Plenderleith LJ, Sundararaman SA, et al. Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. Int J Parasitol. 2017;472:87–97.
Roberts D, Curtis C, Tren R, Sharp B, Shiff C, Bate R. Malaria control and public health. Emerg Infect Dis. 2004;10:1170–1.
WHO. World Malaria report 2015. Geneva: World Health Organization; 2015.
Perkins SL, Austin CC. Four new species of Plasmodium from new guinea lizards: integrating morphology and molecules. J Parasitol. 2009;95:424–43.
Schaer J, Perkins SL, Decherd J, Leendertze FH, Fahr J, Weber N, et al. High diversity of west african bat malaria parasites and a tight link with rodent Plasmodium taxa. Proc Natl Acad Sci USA. 2013;110:17415–9.
Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–45.
Duval L, Robert V, Csorba G, Hassanin A, Randrianarivelojosia M, Walston J, et al. Multiple host-switching of Haemosporidia parasites in bats. Malar J. 2007;6:157.
Witsenburg F, Salamin N, Christe P. The evolutionary host switches of Polychromophilus: a multi-gene phylogeny of the bat malaria genus suggests a second invasion of mammals by a haemosporidian parasite. Malar J. 2012;11:53.
Schaer J, Reeder DM, Vodzak ME, Olival KJ, Weber N, Mayer F, et al. Nycteria parasites of Afrotropical insectivorous bats. Int J Parasitol. 2015;45:375–84.
Lutz HL, Patterson BD, Kerbis Peterhans JC, Stanley WT, Webala PW, Gnoske TP, et al. Diverse sampling of East African haemosporidians reveals chiropteran origin of malaria parasites in primates and rodents. Mol Phylogenet Evol. 2016;99:7–15.
Landau I, Rosin G, Miltgen F, Hugot J-P, Leger N, Beveridge I, et al. Sur le genre Polychromophilus (Haemoproteidae, parasite de Microchiroptères). Ann Parasitol. 1980;55:13–32.
Garnham PC, Lainson R, Shaw JJ. A contribution to the study of the haematozoon parasites of bats. A new mammalian haemoproteid, Polychromophilus deanei n. sp. Mem Inst Oswaldo Cruz. 1971;69:119–27.
Gardner RA, Molyneux DH. Polychromophilus murinus: a malarial parasite of bats: life-history and ultrastructural studies. Parasitol. 1988;96:591–605.
Adam JP, Landau I. Developmental stages of Polychromophilus sp., a parasite of insectivorous bats from the Congo-Brazzaville, in the nycteribiid fly Penicillidia fulvida Bigot 1889. Trans R Soc Trop Med Hyg. 1973;67:5–6.
Landau I, Chavatte JM, Karadjian G, Chabaud A, Beveridge I. The haemosporidian parasites of bats with description of Sprattiella alecto gen. nov., sp. nov. Parasite. 2012;19:137–46.
Duval L, Mejean C, Maganga GD, Makanga BK, Mangama Koumba LB, Peirce MA, et al. The chiropteran haemosporidian Polychromophilus melanipherus: a worldwide species complex restricted to the family Miniopteridae. Infect Genet Evol. 2012;12:1558–66.
Megali A, Yannic G, Christe P. Disease in the dark: molecular characterization of Polychromophilus murinus in temperate zone bats revealed a worldwide distribution of this malaria-like disease. Mol Ecol. 2011;20:1039–48.
Landau I, Chabaud AG. Description of P. cyclopsi n. sp. a parasite of the microchiropteran bat Hipposideros cyclops in Gabon. Ann Parasitol Hum Comp. 1978;53:247–53.
La Dionisi A. malaria di alcune specie di pipistrelli. Atti della Societa per gli Studi della Malaria. 1899;1899:133–75.
Marinkelle CJ. The haemoproteid parasite, Bioccala deanei, from a Colombian bat, Eptesicus fuscus (Vespertilionidae). Ann Trop Med Parasitol. 1995;89:89–91.
Peterson RL, Eger JL, Mitchell L. Chiroptères. Faune de Madagascar. Paris: Muséum national d’Histoire naturelle; 1995.
Goodman SM. Les chauves-souris de Madagascar. Antananarivo: Association Vahatra; 2011.
Goodman SM, Ramasindrazana B, Maminirina CP, Schoeman MC, Appleton B. Morphological, bioacoustical, and genetic variation in Miniopterus bats from eastern Madagascar, with the description of a new species. Zootaxa. 2011;2880:1–19.
Goodman SM, Rakotondramanana CF, Ramasindrazana B, Kearney T, Monadjem A, Schoeman MC, et al. An integrative approach to characterize Malagasy bats of the subfamily Vespertilioninae Gray, 1821, with the description of a new species of Hypsugo. Zool J Linn Soc. 2015;173:988–1018.
Goodman SM, Ramasindrazana B, Naughton K, Appleton B. Description of a new species of the Miniopterus aelleni group (Chiroptera: Miniopteridae) from upland areas of central and northern Madagascar. Zootaxa. 2015;3936:538–58.
Goodman SM, Ramasindrazana B. Bats or the order Chiroptera. In: Goodman SM, Raherilalao MJ, editors. Atlas of selected land vertebrates of Madagascar. Antananarivo: Association Vahatra; 2013. p. 169–209.
Raharimanga V, Ariey F, Cardiff SG, Goodman SM, Tall A, Rousset D, et al. Hémoparasites des chauves-souris à Madagascar. Arch Inst Pasteur Madag. 2003;69:70–6.
Goodman SM, Maminirina CP, Bradman HM, Christidis L, Appleton BR. Patterns of morphological and genetic variation in the endemic Malagasy bat Miniopterus gleni (Chiroptera: Miniopteridae), with the description of a new species, M. griffithsi. J Zool Syst Evol Res. 2010;48:75–86.
Tortosa P, Dsouli N, Gomard Y, Ramasindrazana B, Dick CW, Goodman SM. Evolutionary history of Indian Ocean nycteribiid bat flies mirroring the ecology of their hosts. PLoS ONE. 2013;8:e75215.
Lagadec E, Gomard Y, Guernier V, Dietrich M, Pascalis H, Temmam S, et al. Pathogenic Leptospira spp. in bats, Madagascar and Union of the Comoros. Emerg Infect Dis. 2012;18:1696–8.
Gomard Y, Dietrich M, Wieseke N, Ramasindrazana B, Lagadec E, Goodman SM, et al. Malagasy bats shelter a considerable genetic diversity of pathogenic Leptospira suggesting notable host-specificity patterns. FEMS Microbiol Ecol. 2016;92:fiw037.
Ramasindrazana B, Goodman SM, Gomard Y, Dick CW, Tortosa P. Hidden diversity of Nycteribiidae (Diptera) bat flies from the Malagasy Region and insights on host parasite interactions. Parasit Vectors. 2017;10:630.
Dick CW, Patterson BD. Bat flies—obligate ectoparasites of bats. In: Morand S, Krasnov BR, Poulin R, editors. Micromammals and macroparasites: from evolutionary ecology to management. Japan: Springer; 2006. p. 179–94.
Wilkinson DA, Temmam S, Lebarbenchon C, Lagadec E, Chotte J, Guillebaud J, et al. Identification of novel paramyxoviruses in insectivorous bats of the southwest Indian Ocean. Virus Res. 2012;170:159–63.
Wilkinson DA, Mélade J, Dietrich M, Ramasindrazana B, Soarimalala V, Lagadec E, et al. Highly diverse Morbillivirus-related paramyxoviruses in the wild fauna of southwestern Indian Ocean islands: evidence of exchange between introduced and endemic mammals. J Virol. 2014;88:8268–77.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9.
Paradis E. Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–20.
R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna; 2013. http://www.R-project.org.
Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–6.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772.
Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42.
Valkiunas G, Iezhova TA, Krizanauskiene A, Palinauskas V, Sehgal RNM, Bensch S. A comparative analysis of microscopy and PCR-based detection methods for blood parasites. J Parasitol. 2008;94:1395–401.
Witsenburg F, Schneider F, Christe P. Epidemiological traits of the malaria-like parasite Polychromophilus murinus in the Daubenton’s bat Myotis daubentonii. Parasit Vectors. 2014;7:556.
Bastien M, Jaeger A, Corre ML, Tortosa P, Lebarbenchon C. Haemoproteus iwa in great frigatebirds (Fregata minor) in the islands of the western Indian Ocean. PLoS ONE. 2014;9:e97185.
Sehgal RNM, Buermann W, Harrigan RJ, Bonneaud C, Loiseau C, Chasar A, et al. Spatially explicit predictions of blood parasites in a widely distributed African rainforest bird. Proc R Soc B. 2011;278:1025–33.
Lainson R, Naiff RD. Haemoproteus (Apicomplexa: Haemoproteidae) of tortoises and turtles. Proc R Soc Lond B. 1998;265:941–9.
Witsenburg F, Clément L, López-Baucells A, Palmeirim JM, Pavlinic I, Scaravelli D, et al. How a haemosporidian parasite of bats gets around: the genetic structure of a parasite, vector and host compared. Mol Ecol. 2015;24:926–40.
Stadelmann B, Jacobs DS, Schoeman C, Ruedi M. Phylogeny of African Myotis bats (Chiroptera, Vespertilionidae) inferred from cytochrome b sequences. Acta Chiropt. 2004;6:177–92.
Mer GG, Goldblum N. A haemosporidian of bats. Nature. 1947;159:44.
Jl Obame-Nkoghe, Rahola N, Bourgarel M, Yangari P, Prugnolle F, Maganga GD, et al. Bat flies (Diptera: Nycteribiidae and Streblidae) infesting cave-dwelling bats in Gabon: diversity, dynamics and potential role in Polychromophilus melanipherus transmission. Parasit Vectors. 2016;9:333.
Sikes RS, Gannon WL, The animal care and use committee of the american society of mammalogists. Guidelines of the american society of mammalogists for the use of wild mammals in research. J Mammal. 2011;92:235–53.
Authors’ contributions
BR, SMG, PT, and KD designed the study; BR, SMG, EL, ND, and YG performed the sampling; BR carried out the molecular and statistical analyses. BR, PT, and SMG principally wrote the manuscript, which was commented on by all authors. All authors read and approved the final manuscript.
Acknowledgements
We are grateful to the Mention Zoologie et Biodiversité Animale, Université d’Antananarivo, Madagascar National Parks, and the Ministère de l’Environnement et des Forêts of Madagascar for their valuable assistance regarding different administrative aspects and for providing research permits to conduct this work. We would like to thank Claude Fabienne Rakotondramanana and Tsibara Mbohoahy for their assistance in bat sampling. We are also grateful to Julien Mélade, Claire Bernard, and Mbola H. Rakotondratsimba for their assistance and valuable advice on different aspects. We would like to thank the anonymous reviewers for their comments on a previous version of this manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated during this study are included in this manuscript and its additional file.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The capture of bats was conducted in strict accordance with the terms of research permits issued by Malagasy authorities (Direction du Système des Aires Protégées, Direction Générale de l’Environnement et des Forêts, and Madagascar National Parks) and following national laws (Permit Numbers: 194/12/MEF/SG/DGF/DCB.SAP/SCB, 283/11/MEF/SG/DGF/DCB.SAP/SCB; 067/12/MEF/SG/DGF/DCB.SAP/SCBSE; 036/12/MEF/SG/DGF/DCB.SAP/SCBSE, and N° 077/12/MEF/SG/DGF/DCB.SAP/SCBSE). Bats were trapped, manipulated, and euthanized in strict accordance with guidelines accepted by these different national authorities and the scientific community for the handling of wild animals [52]. A CITES permit from the Malagasy authority was issued for the exportation of tissue samples of Pteropus rufus to La Réunion (permit 243C-EA06/MG12).
Funding
Financial supports to conduct this work were graciously provided by the Fonds Européen de Développement Régional FEDER POCT Réunion (Pathogènes associés à la faune sauvage Océan Indien #31189) and the Volkswagen Foundation. BR received postdoctoral fellowship from RunEmerge project funded by the European Frame work programme FP7 Capacities/Regpot, a post-doctoral grants from “Fonds de Coopération Régionale” of the Préfecture de La Réunion, and from the Dr. Ralph and Marian Falk Medical Research Trust to The Field Museum of Natural History, Chicago.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Table S1.
Parasites included in the present study, including Haplotype, Isolate, GenBank accession numbers, host species, Museum voucher and origin. Molecular data produced in the frame of the present work are marked with an asterisk (*). FMNH = Field Museum of Natural History, UADBA = Université d’Antananarivo, Département de Biologie Animale, NA: not available.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ramasindrazana, B., Goodman, S.M., Dsouli, N. et al. Polychromophilus spp. (Haemosporida) in Malagasy bats: host specificity and insights on invertebrate vectors. Malar J 17, 318 (2018). https://doi.org/10.1186/s12936-018-2461-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-018-2461-8