Abstract
Background
Protein regulator of cytokinesis 1 (PRC1) has been reported to play important role in the pathogenesis of various cancers. However, its role in colon cancer has not been studied. Here, we aimed to investigate the biological functions and potential mechanism of PRC1 in colon cancer.
Methods
The expression level of PRC1 in colon cancer tissues and cell lines was detected by quantitative real-time polymerase chain reaction (qRT-PCR), Western blotting, and immunohistochemical (IHC) staining of a tissue microarray (TMA). Furthermore, colon cancer cell lines HCT116 and SW480 were treated with short hairpin RNAs against PRC1. The biological function of PRC1 was determined by MTT proliferation, colony formation assay, cell cycle, and apoptosis assays. Then, an in vivo tumor formation assay was conducted to explore the effects of PRC1 on tumor growth.
Results
The mRNA and protein expression levels of PRC1 were highly expressed in colon cancer tissues and cell lines. PRC1 expression was associated with clinicopathological characteristics and overall survival of patients with colon cancer. Knockdown of PRC1 could decrease proliferation and colony forming ability of colon cancer cells, as well as arrested more cells at G2/M phase and promoted cell apoptosis. In cancer cells, the expression pattern of protein regulators included in cell cycle and apoptosis progress were reverted by PRC1 down-regulation. Additionally, PRC1 down-regulation could suppress colon tumor growth and differentiation.
Conclusions
We confirmed that PRC1 was overexpressed in colon cancer and was associated with poor prognosis of colon cancer patients. PRC1 down-regulation could arrest cell cycle at G2/M stage, inhibit proliferation, and elicit apoptosis. These findings showed the potential of PRC1 to be used for therapeutic approaches in colon cancer.
Similar content being viewed by others
Background
Colon cancer is the third most common cancer, accounting for around 10% of all diagnosed cancers worldwide [1]. Although nationwide screening programs cause an increase in colon cancer incidence, lifestyle [2], heredity [3] and environmental factors [4] might be attributable to the increased the incidence. However, the reasons of this increase are not exactly understood. Patients with colon cancer display a wide range of symptoms, such as abdominal pain, anaemia, disorders of bowel habits, and rectal bleeding. For diagnosis of colon cancer, colonoscopy and computed tomography colonography are increasingly used, with histology as identification [5]. Moreover, researchers have confirmed the potential of identifying patients with colon cancer according to gene expression signatures [6, 7]. Accordingly, gene-based therapy may be useful to eradicate colon cancer, which has been proved as a promising therapeutic strategy [8, 9]. Here, we investigated whether protein regulator of cytokinesis (PRC1) might function as the diagnosis and prognosis of colon and whether showed a potential in targeted treatments of colon cancer.
PRC1 is composed of 620 amino acids, which is structured into Lys/Arg-rich, MT-binding, dimerization, and rod domains [10,11,12]. PRC1 variably resides in different types of tissues and tightly affects cell division [13]. PRC1 copy number changes and missense mutations disrupt the tight spatiotemporal regulation of cytokinesis [14]. The deregulation of PRC1 contributes to tumorigenesis and cancer process [14, 15]. A recent study confirmed that the dysregulation of PRC1 might be responsible for biochemical recurrence in patients with prostate cancer, implying that PRC1 could be considered as a prognostic indicator [16]. Consistently, a previous report revealed PRC1 was accumulated in the breast cancer, and PRC1 promoter exhibited cancer-specific activity [17]. Besides, evidence suggests that PRC1 is involved in breast tumorigenesis, and it is a promising target for clinical treatment of breast cancer [16]. However, it is still unclear whether PRC1 shows similar expression profile and what roles PRC1 exactly plays in colon cancer.
PRC1 was reported as the second highest ranking gene studied to be correlated with a high grade of chromosomal instability in diverse tumors [18]. Its overexpression is associated with enhanced aneuploidy which leads to worse patient outcome [14, 15, 19]. Genetic abnormalities may cause the deregulation of PRC1 since genetic events such as missense mutations, deletions and amplifications are circularly determined by genomic profiling [20, 21]. Besides, PRC1 is deregulated by signaling pathways at transcriptional level, such as Wnt, p53 and non-estrogen receptor in various cancer, indicating that signaling transduction cascades may be therapeutically utilized to govern rates of cytokinesis defects [22,23,24]. Consistently, comprehensive analysis of its deregulation is conducive to develop new therapeutic candidates for colon cancer.
Given these confirmations of PRC1 functioning in cancer disease, it is unclear whether PRC1 operates in colon cancer. In this study, we sought to analyze the expression profile of PRC1 in colon cancer as well as the potential affinity between PRC1 and cancer prognosis. Next, we knocked down PRC1 to study its biological function in colon cancer. Our findings demonstrated the involvement of PRC1 in colon cancer, and highlighted its potential as a diagnostic, therapeutic, and prognostic maker for colon cancer.
Methods
Patients and tissue samples
A total of 40 patients (20 males and 20 females) ranged 41–85 years (mean, 68.1 years) undergoing surgical resection of colon carcinoma at the Center of Tumor, Inner Mongolia People’s Hospital (Inner Mongolia Autonomous Region, China) between July 2014 and March 2015 were selected for this study. The clinical characteristics of included patients are shown in Additional file 1: Table S1. The colon carcinoma tissues and paired adjacent normal tissues were obtained from surgeries. None of patients received treatment prior to surgery, such as radiotherapy, chemotherapy, biotherapy, or other combination therapies. All samples were confirmed by histopathology as colon carcinoma or adjacent normal colon tissue. Then, tissues were frozen quickly in liquid nitrogen. All patients included in this study had signed written informed consents. This study obtained permission from the Research Ethics Committee of Inner Mongolia People’s Hospital.
Bioinformatics analysis
The mRNA expressions of PRC1 in colon and normal tissues were obtained from Oncomine (https://www.oncom ine.org/resou rce/login.html) as follows: Sabates-Bellver Colon [25], Hong Colorectal [26], and Skrzypczak Colorectal [27].
Immunohistochemistry (IHC) and scoring
IHC was performed on a tissue microarray (TMA) slide. The colon cancer TMA (Cat: HCol-Ade180Sur-08) purchased by the Shanghai Outdo Biotech Co., Ltd (Shanghai. China) was chosen for evaluating PRC1 expression. This TMA contained 90 patients with colon cancer who underwent surgical resection between July 2004 and June 2009. The final followed up date was August 2015 and the follow-up time was 5–10 years. The mean age of the patients was 67.7 years (range, 47–90 years), including 45 males and 45 females. Meanwhile, the clinical characteristics of these patients are described in Additional file 1: Table S2. This microarray contained colon cancer tissues and paired normal adjacent tissues from 90 patients. TMA slide was cut into 4-μm-thick section and then were deparaffinized. Antigen retrieval was performed by microwaving in citric acid (pH 6.0) for 5 min, followed by incubation with the primary antibody against PRC1 (51248, Abcam, Cambridge, UK) at 4 °C overnight. After incubation with secondary antibody, images were collected and evaluated blindly by two independent pathologists. The two-way scoring system was used for staining quantification. A semi-quantitative method was resorted to assess the staining intensity with a four point scale: no staining (0), weak staining (1), moderate staining (2), and strong staining (3). Meanwhile, the proportion of positively stained cells was determined as following: 0% (0), 1–25% (1), 26–50% (2), 51–75% (3), and 76–100% (4). The final expression level of protein, ranging from 0 to 12, was calculated by multiplying the intensity score and the proportion score. The average immunohistochemistry score of PRC1 was used as a cut-off to divide the cases into low expression group and high expression group.
Cell culture
Human colon cancer cells lines (HCT116, SW480, Caco-2, and HT-29) and normal human colon epithelial lines (CCD-18Co) were provided by the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). HCT116 cells were cultured in McCOY’s 5A (GIBCO-BRL, Invitrogen, Carlsbad, CA, USA); and CCD-18Co, SW480, Caco-2, and HT-29 cells were cultured in RPMI-1640 (Gibco, Carlsbad, CA, USA). All the culture media were supplemented with 10% fetal bovine serum (FBS). Meanwhile, all cells were cultured in a humidified incubator under standard culture condition (5% CO2, 37 °C).
Short hairpin RNA (shRNA) transfection of colon cancer cell line SW480, HT-29 and HCT116
Two human PRC1 shRNA (shPRC1#1 5′-CCTGAAGGAAAGACTCATCAA-3′ and shPRC1#2 5′-CAGGAACATTCAAAGGCATTT-3′) and control-shRNA were synthesized by GenePharma (Shanghai, China). shPRC1-1 and shPRC1-2 were inserted into pcDNA3.1-PRC1 vectors (Genechem, Shanghai, China) for specifically targeting PRC1 mRNA. For the control lentiviral, shControl sequence (did not target any known gene) was inserted into pcDNA3.1 (Addgene). The construction of the recombinant plasmid was resorted to GenePharma (Shanghai, China). Viruses were kept in the PEG-it virus reagent (System Biosciences, Piscataway, NJ, USA) and maintained at -80 °C. When cells (SW480, HT-29, and HCT116) reached 50–60% confluence, the constructed shRNAs and shControl were transfected into cells using Lipofectamine 2000 (Invitrogen, Life Technologies) according to the manufacturer’s instructions. Successful knockdown of PRC1 expression was tested by quantitative real-time polymerase chain reaction (qRT-PCR) and western blotting.
RNA extraction and qRT-PCR
Total RNA was extracted from frozen tissues and cultured cells using TRIzol Kit (Invitrogen) based on the manufacturer’s instruction. The complementary DNA (cDNA) was reversely transcribed using the SuperScript III First-Strand Kit (Invitrogen). qRT-PCR reaction was conducted on the ABI7500 Real-Time PCR (Applied Biosystems, Foster City, CA, USA) by using SYBR Premix Ex Taq II (Takara, Dalian, China). GAPDH was used for a reference gene. The relative expression of PRC1 was quantified using 2−△△CT method. All experiments were performed in triplicate. The Primer sequences were as follows (synthesized by Sangon Biotech (Shanghai, China)): PRC1 forward, 5′-TAGACCACACCCCAGACACA-3′ and reverse, 5′-GTGGCCACAGCTTCTCTTTCP-3′; and GAPDH forward, 5′-GCAAATTCCATGGCACCGT-3′, and reverse, 5′-GCCCCACTTGATTTTGGAGG-3′.
Western blotting assay
The expression levels of PRC1 in cultured cell lines and tumor tissues were detected by western blotting. Total proteins from cells and tissues were extracted and lysed by RIPA buffer (Beyotime, Beijing, China) containing protease and phosphatase inhibitors (Roche, Basel, Switzerland). The concentration of extracted protein was examined using Bio-Rad Protein Assay kit (Bio-Rad, Hercules, CA, USA). Protein extracts (30 μg) were separated on 10–12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA). Commercial primary antibodies including PRC1 (santa cruz; #8356; 1:1000 dilution), GAPDH (abcam; #ab181602; 1:10000), cyclin B1 (CST; #4138; 1:1000), cyclin dependent kinase 1 (Cdc2; CST; #9116; 1:1000), P21 (Santa cruz; #397; 1:1000), cell division cycle 25c (Cdc25c; CST; #4688; 1:1000), and P27 (CST; #3686; 1:1000) were incubated with the PVDF membrane overnight at 4 °C. The primary antibodies were then probed using the appropriate secondary antibodies at room temperature for 1 h, followed by ECL Western blotting Detection System (Amersham, Piscataway, NJ, USA) for detecting protein signal band. GAPDH served as an endogenous reference and each experiment was run in triplicate. The results of western blotting bands were quantified using Image J software (Rawak Software Inc, Stuttgart, Germany).
Cell viability assay
The effect of PRC1 on the viability of colon cancer cell lines was determined by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) (Sigma-Aldrich) assay as described previously [28]. Briefly, shPRC1 (shPRC1-1 and shPRC1-2) and shControl were used to transfect the colon cancer cells (SW480, HT-29, and HCT116). After lentiviral transduction, the cells were seeded at 2.5 × 104 cells/well in 96-well plates, followed by incubation at 37 °C in an atmosphere containing 5% CO2 for 7 days. From day 3 to 7, 100 mL of MTT (5 mg/mL) was added and incubated for 4 h each day. Afterwards, the entire supernatant was discarded and then 150 μL dimethyl sulfoxide was added to each well to completely dissolve the formazan. The plates were lightly shaken on the table for 10 min, and optical density values were measured at a wavelength of 490 nm using a spectrophotometer microplate reader (BioTek, Winooski, VT, USA). All analysis was done three times in triplicate.
Colony formation assay
Transfected SW480, HT-29, and HCT116 cells (1000 cells/well) were seeded into 6-well plates with fresh medium containing 10% FBS. Cells were cultured for 14 days, and the medium was changed every 4 days. Subsequently, the cells were fixed with methanol for 15 min and stained with 1% crystal violet (Solarbio) for 20 min at room temperature. The cells were dried and images were captured using a microscope (Olympus, FV500-IX71, Japan) to count the number of colonies containing > 50 cells.
Cell cycle analysis
Cell cycle analysis was conducted to verify whether PRC1 regulated the cell cycle. In short, the SW480 and HCT116 cells were transfected with shPRC1 (shPRC1-1 and shPRC1-2) and shControl for 5 days. Then, transfected cells were harvested and fixed with 70% ice-cold ethanol overnight at 4 °C. Cell suspensions were stained with 50 μg/mL of propidium iodide (PI, containing 100 μg/mL RNAase) and analyzed using a flow cytometry (BD Biosciences, San Diego, CA, USA). The experimental procedures were carried out three times in triplicates.
Apoptosis assay
Flow cytometry was used to detect the apoptotic rate. Colon cells (SW480 and HCT116) transfected with shPRC1 (shPRC1-1 and shPRC1-2) or shControl were plated into 6-well plates and grown to 90% confluence. Following incubation, cell suspensions were harvested and incubated with 10 μL Annexin V-FITC (Beijing Biosea Biotechnology co., LTD., Beijing, China) staining for 10–15 min at room temperature avoiding lights. Then, the cells were analyzed using a flow cytometer (BD Biosciences, San Diego, CA, USA).
Mice experiments
Four-week old male athymic BALB/c nude mice were provided by SLAC Laboratory Animal Co. (Shanghai, China). The experimental protocols were permitted by the Institutional Animal Care and Use Committee of Jinling Hospital. Mice were kept in a specific pathogen-free environment and randomly divided into two groups (7 mice per group). Xenograft tumor models were established by subcutaneously injecting of stable shControl- or shPRC1-1-expressing HCT116 cells (3 × 107) into the left flank of nude mice. The volume and weight of the tumors were measured every three days for a total of three weeks. Tumor volume (V, mm3) was measured using calipers and calculated according to the formula: V = length × width2 × 0.5. After 21 days, mice were euthanized and tumors were extracted for further analysis. These tissues were fixed in 4% paraformaldehyde for 24 h. The tissues were processed for hematoxylin and eosin (H&E) staining as well as immunohistochemical staining for Cdc2, Cyclin B1, ki67, and PRC1. The primary antibody used in this analysis was as follows: PRC1 (abcam; #51248: 1:500), Cyclin B1 (CST; #4138; 1:1000), Cdc2 (CST; #9116; 1:1000), and Ki67 (CST; #9449; 1:400). The images were obtained under a brightfield microscopy (Olympus, Tokyo, Japan).
Statistical analysis
All data were analyzed using GraphPad Prism 6 software (GraphPad, San Diego, CA, USA) and shown as the mean ± standard deviation (SD). Differences between two groups were analyzed using the t-test, and comparisons among multiple groups were applied by one-way analysis of variance (ANOVA). Chi-square test was used to assess the association between PRC1 level and clinical characteristics of colon cancer patients. The cut-off value to divide patients into high and low expression groups was defined as the average expression of PRC1. Kaplan–Meier method was used to plot survival curves and log-rank test was utilized to compare difference. Cox proportional hazards model was chosen to evaluate independent prognostic factor. P < 0.05 was considered statistically significant.
Results
PRC1 was high expression in colon cancer tissues and colon cell lines
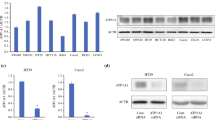
First, we investigated the PRC1 expression in datasets available from Oncomine (https://www.oncomine.org). The data of Sabates-Bellver colon, Hong colorectal, and Skrzypczak colorectal showed that PRC1 was overexpressed in colon cancer compared to normal tissues (Fig. 1a). Then, PRC1 expression level was examined in 40 colon cancer tissues and paired adjacent normal tissues by qRT-PCR. The results indicated that PRC1 expression was significantly higher in cancer tissues than that in normal tissues (P < 0.001, Fig. 1b). Moreover, we selected 12 pairs of colon tumor tissues and adjacent normal tissues to detect the expression of PRC1 protein. Compared with the normal tissues, the PRC1 protein expression level was significantly elevated in colon cancer tissues (P < 0.001, Fig. 1c). Simultaneously, we examined PRC1 in cell lines by qRT-PCR and western blotting. The PRC1 level was obviously upregulated in colon cell lines (HCT116, SW480, Caco-2, and HT-29) compared with the human colon epithelial line CCD-18Co (P < 0.05, Fig. 1d, e).
PRC1 was overexpressed in colon cancer tissues and cells. a Oncomine data showed that PRC1was highly expressed in colon tissues compared with normal tissues. b The mRNA expression level of PRC1 was significantly up-regulated in colon cancer tissues compared with adjacent normal tissues in 40 paired tissues. c The PRC1 protein expression level was significantly elevated in colon cancer tissues compared with adjacent normal tissues in selected 12 paired tissues. d, e The expression level of PRC1 was higher in the colon cancer cell lines (HCT116, SW480, Caco-2, and HT-29) than in the normal human epithelial line (CCD-18Co) on mRNA (d) and protein level (e). Data were presented as the mean ± standard deviation (SD). #P < 0.001 and *P < 0.05
PRC1 was associated with clinicopathlogical parameters and poor prognosis in colon cancer
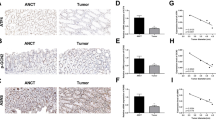
In order to further explore the expression pattern of PRC1 in colon cancer, we conducted IHC in an established TMA, which composed of 90 paired colon cancer tissues and neighboring non-cancerous tissues. As shown in Fig. 2a, results from TMA suggested that the protein level of PRC1 was universally higher expressed in colon cancer tissues relative to the adjacent normal tissues (P < 0.001). In addition, the 90 patients could be grouped into low-expression and high-expression groups according to the immunohistochemical score of PRC1, using 6.2 as a cut-off value (Fig. 2b). The relationship between PRC1 expression and clinicopathlogical parameters of patients with colon cancer is shown in Table 1. The results demonstrated that high expression of PRC1 was associated with high TNM stage (P < 0.01), large tumor size (> 5 cm, P < 0.05), and lymph node metastasis (P < 0.05); while there was no relationship between PRC1 expression and gender (P = 0.673) as well as age (P = 0.539).
PRC1 was associated with poor prognosis in colon cancer. a IHC images of PRC1 protein in 90 colon cancer tissues and adjacent normal tissues obtained from TMA. The PRC1 protein expression score in tumor tissues was significantly higher than normal tissues. b Representative images of PRC1 IHC staining from colon cancer TMA. Left: high PRC1 expression in colon cancer (≥ 6.2), right: low PRC1 expression in colon cancer (< 6.2). c OS was poor in patients with high expression of PRC1 than in those with low expression among 90 colon cancer patients. IHC immunohistochemistry, TMA tissue microarray, OS overall survival
Kaplan–Meier survival analysis revealed that colon cancer patients with high expression level of PRC1 had a poorer overall survival (OS, Fig. 2c). Additionally, cox regression analysis confirmed that PRC1 was an independent prognostic factor for colon cancer (95% CI: 1.840–12.346, P < 0.001). Moreover, results showed that lymph node metastasis, p-TNM stages and PRC1 expression were significantly associated with OS (all P < 0.05, Table 2).
Silencing of PRC1 inhibited cell viability and colony forming ability of colon cancer cells
To explore the biological function of PRC1 in colon cancer cells, we established the colon cancer cells (SW480, HT-29, and HCT116) with lentivirus mediated PRC1 knockdown, which was validated by qRT-PCR and western blotting. The results confirmed that both mRNA and protein expression levels of PRC1 were significantly suppressed in sh-PRC1 transfected cell lines (P < 0.05) (Fig. 3a, b). MTT and colony forming assays were performed to determine the influence of PRC1 on cell proliferation. As shown in Fig. 4a, cell proliferation was significantly inhibited in the shPRC1 groups (shPRC1-1 and shPRC1-2) compared with that in shControl group in SW480, HT-29, and HCT116 cells (P < 0.01). In addition, colony formation assay was conducted to evaluate the colony formation ability of colon cells. As presented in Fig. 4b, the decreased number of colonies was observed in the cells transfected with shPRC1 (shPRC1-1 and shPRC1-2) (P < 0.01).
The interfering efficiency of two shRNA targeting PRC1 (shPRC1-1 and shPRC1-2) was exaimed by qRT-PCR (a) and western blotting (b). Compared with normal group (shControl), the expression levels of PRC1 in shPRC1 transfected colon cancer cell lines (SW480, HT-29, and HCT116) were significantly decreased. **P < 0.01
Silencing of PRC1 inhibited cell viability and colony forming action ability of colon cancer cells. a The cell grow rate of colon cancer cells (SW480, HT-29, and HCT116) was decreased in shPRC1 transfected groups (shPRC1-1 and shPRC1-2). b PRC1 knockdown inhibited SW480, HT-29, and HCT116 cells proliferation. **P < 0.01
Silencing of PRC1 arrested more cells in G2/M phase and promoted apoptosis of colon cancer cells
Cell cycle distribution of SW480 and HCT116 cells in each group (shControl, shPRC1-1, and shPRC1-2) was evaluated by PI staining. As shown in Fig. 5a, b, silencing of PRC1 expression in HCT116 and SW480 cells, compared with the shControl group, led to a significant increase in G2/M phase arrest and a decrease in the G0/G1 cell population (P < 0.01). To further elucidate molecular mechanisms of PRC1 regulating cell proliferation, we detected the cell cycle-related proteins levels in HCT116 and SW480 cells by western blotting. Compared with shControl group, cell cycle inhibitors at G1 phase (p21 and p27) were significantly up-regulated (P < 0.05), while cell cycle regulatory proteins (Cdc25c, Cdc2, and Cyclin B1) were down-regulated in the cells transfected with shPRC1-1 and shPRC1-2 (P < 0.05, Fig. 5c). Moreover, flow cytometry was performed for detection of cell apoptosis of HCT116 and SW480 cells. Compared with the shControl group, the apoptosis rate was increased in the shPRC1 transfected cells (P < 0.01) (Fig. 5d, e). Furthermore, the expression levels of apoptosis-related proteins were detected by western blotting. Our results showed that PRC1 positively regulated cleaved caspase 3 and Bax, as well as negatively regulated full length PARP, Bcl2 and Bcl-xl (P < 0.05) (Fig. 5f). These results provided the evidences that PRC1 showed oncogenic effects via regulating G2/M cell cycle and cell apoptosis.
Silencing of PRC1 arrested more cells in G2/M phase and induces cell apoptosis in HCT116 and SW480 cells. a Cell cycle distribution of HCT116 and SW480 cells in the three groups (shControl, shPRC1-1, and shPRC1-2). b Percentage of cells in cell cycle phase G0/GA, S, and G2/M. c The expression levels of cell cycle related proteins (Cdc25c, Cdc2, Cyclin B1, P21, and P27) were detected by western blotting. d Cell apoptosis in HCT116 and SW480 cells detected by flow cytometry. e Apoptosis rate in HCT116 and SW480 cells in the three groups. f The expression level apoptosis related proteins (pro caspase 3, cleaved caspase 3, full length PARP, Bcl2, Bcl-xl, and Bax) was detected by western blotting. Data were expressed as the mean ± standard deviation (mean ± SD). *P < 0.05, **P < 0.01, # P < 0.001
Silencing of PRC1 mitigated the growth of tumors
Finally, we established xenograft tumor model to investigate the effect of PRC1 on tumor growth in vivo. Results suggested that the tumors of the PRC1 knockdown group (shPRC1-1) were significantly smaller and lighter than those of the control group (P < 0.01) (Fig. 6a–c). Subsequently, immunohistochemical staining of xenograft tissues collected from nude mice demonstrated that knockdown of PRC1 could significantly inhibit the expression of cancer differentiation-related proteins (Cdc2, Cyclin B1, and ki67) (P < 0.05, Fig. 6d). Overall, these findings illustrated that inhibition of PRC1 could suppress colon tumor growth and differentiation.
Knockdown of PRC1 inhibited tumor growth in vivo. a Xenograft tumor formation of HCT116 cells transfected shControl and shPRC1-1. b, c The volume and weight of tumor were calculated. d The resected tumors were subjected to H&E staining and IHC staining for Cdc2, cyclin B1, Ki67 and PRC1 (× 200 magnification). H&E hematoxylin–eosin, IHC immunohistochemical, AO average optical. Data were expressed as the mean ± standard deviation (mean ± SD). **P < 0.01
Discussion
In this study, we observed that PRC1 was significantly up-regulated in colon tissues and cell lines. The expression level of PRC1 was associated with several clinicophathological characteristics in colon cancer patients, including tumor size, lymph node metastasis, and p-TNM stages; in addition, patients with high expression of PRC1 suggested unfavorable prognosis, which indicated that PRC1 might serve an important role in the development and progression of colon cancer. Furthermore, the biological function of PRC1 was investigated in vitro and in vivo, and results showed that knockdown of PRC1 inhibited proliferation and promoted apoptosis of colon cancer cells. Meanwhile, silencing of PRC1 resulted in more cells arrested at G2/M phase. The results of mice experiment demonstrated that knockdown of PRC1 could inhibit tumor growth. Our data provided more evidence that PRC1 might serve a therapeutic target for colon cancer.
PRC1 is involved in the completion of cytokinesis and is abnormally regulated in a tumor-specific manner. Numbers studies have reported the role of PRC1 in cancer development. Luo et al. found that PRC1 expression levels were significantly higher in prostate cancer (PCa) tissues than those in normal tissues; in addition, overexpression of PRC1 was significantly associated with advanced clinicopathologic features and a shorter biochemical recurrence-free survival in patients with PCa [29]. Bu et al. observed that the mRNA and protein expression levels were up-regulated in high-grade serous ovarian carcinoma (HGSOC) tissues, and knockdown of PRC1 could reduce the proliferation, metastasis, and in vitro multidrug resistance of ovarian cancer cell [30]. Consistent with previous studies, our experiments also indicated that PRC1 was obviously higher in tumor tissues compared with normal paracancer tissues. Meanwhile, its expression level was associated with the clinical indicators and OS of patients with colon cancer, revealing that PRC1 had carcinogenic effect and might be an ideal target for the therapy of colon cancer.
Most notably, our findings showed that silencing of PRC1 could significantly inhibit the proliferation and attenuate the colony forming ability of colon cancer cells. Proliferation is an important feature of life activities and its manifestation at the cellular level is cell division [31]. The accurate entry of cells into the growth and division cycle is a prerequisite for maintaining normal cell proliferation and genome stability. However, the tetraploid and chromosomal instability caused by the failure of accurate cell division can promote the occurrence and development of tumors [32]. Li et al. suggested that PRC1 was essential for cytokinesis and normal cell cleavage, and deregulation of PRC1 protein could cause cell division defects, thereby promoting chromosomal instability, leading to tumor heterogeneity and tumor evolution [33]. Thus, we speculated that abnormal expression of PRC1 led to erroneous cell division, which in turn promoted the development of colon cancer. Subsequently, we explored the effect of PRC1 on the cell cycle and apoptosis of colon cancer cells.
Our results showed that silencing of PRC1 expression arrested cell cycle progress at the G2/M interphase. Meanwhile, the expression levels of cell cycle related proteins were detected by western blotting. Silencing of PRC1 leaded to decrease in the protein expression level of Cdc25c, Cdc2, and Cyclin B1. Previous study revealed that Cdc25c-mediated de-phosphorylation of apoptosis signal-regulating kinase 1 (ASK1) and Cdc25 overexpression blocked ASK1-mediated apoptosis in a cell cycle-dependent manner, showing that Cdc25c functioned in G2/M checkpoint-mediated apoptosis [34]. Furthermore, cyclin B1 interacting through its P-box with Cdc25 activated Cdc25c and subsequently activated Cdc2 kinase [35]. Meanwhile, we noticed that inhibition of PRC1 resulted in increases of P21 and P27. As reported, P21 and P27, possessing the overlapping functions with each other, were involved in the cell cycle exit at G1-phase [36]. What’s more, down-regulation of PRC1 was responsible for the cleavage of caspase 3 and PARP, increased the Bax expression, as well as decreased the Bcl2 and Bcl-xl expression, which led to cell apoptosis [37]. It was therefore probable that PRC1 decrease arrested cell cycle at the G2/M interphase and caused apoptosis through affecting regulator expression. However, how PRC1 affects their expression remains to be determined.
Finally, our findings from xenograft tumor growth assays demonstrated that silencing of PRC1 expression inhibited the weight and volume of the tumor. This inhibition of PRC1 expression resulted in decreased the protein expression levels of Cdc2 and Cyclin B1 in xenograft tumors, which was in consistence with the results from in vitro experiments. Moreover, another importance observation in our studies was that PRC1 down-regulation decreased Ki67 expression. Of note, Ki67 is exclusively expressed within the cell nucleus during interphase while relocates on the chromosomal surface in mitosis, being strikingly associated with proliferative activities [38, 39]. Our findings strongly suggested that PRC1 down-regulation inhibited xenograft-tumor growth in vivo.
To the best of our knowledge, this is the first study to explore the biological function of PRC1 in the pathogenesis of colon cancer. Nevertheless, our study has several limitations. Due to the incomplete follow-up data for 5–10 years, the association between PRC1 expression and clinical features (such as OS, gender, age, size tumor, and so on) has not been investigated in the collected 40 patients. Moreover, the precise pathway through which PRC1 influences colon cancer progression has not been explored. In the subsequent research, we will further reveal the potential regulation pathways for PRC1 in colon cancer.
Conclusions
The present study depicted the correlation of colon cancer progress to PRC1 overexpression. Our data presented in this study showed that PRC1 was overexpressed in colon cancer and was associated with poor OS. PRC1 knockdown could arrest cell cycle at G2/M stage, inhibit proliferation, and promote apoptosis. These results indicated the potential of PRC1 to be used for therapeutic approaches in colon cancer.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- PRC1:
-
Protein regulator of cytokinesis 1
- PI:
-
Propidium iodide
- PVDF:
-
Polyvinylidene fluoride
- SD:
-
Standard deviation
- ASK1:
-
Apoptosis signal-regulating kinase 1
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Ouakrim D, Pizot C, Boniol M, Malvezzi M, Boniol M, Negri E, Bota M, Jenkins M, Bleiberg H, Autier P. Trends in colorectal cancer mortality in Europe: retrospective analysis of the WHO mortality database. BMJ. 2015;351:h4970.
Jiao S, Peters U, Berndt S, Brenner H, Butterbach K, Caan B, Carlson C, Chan A, Chang-Claude J, Chanock S, et al. Estimating the heritability of colorectal cancer. Hum Mol Genet. 2014;23(14):3898–905.
Yang C, Wang X, Huang CH, Yuan WJ, Chen ZH. Passive smoking and risk of colorectal cancer: a meta-analysis of observational studies. Asia Pac J Public Health. 2016;28(5):394–403.
Moreno C, Mittal P, Sullivan P, Rutherford R, Staley C, Cardona K, Hawk N, Dixon W, Kitajima H, Kang J, et al. Colorectal cancer initial diagnosis: screening colonoscopy, diagnostic colonoscopy or emergent surgery and tumor stage and size at initial presentation. Clin Colorectal Cancer. 2015;15(1):67–73.
Chen J, Ding J, Wang Z, Zhu J, Wang X, Du J. Identification of downstream metastasis-associated target genes regulated by LSD1 in colon cancer cells. Oncotarget. 2017;8(12):19609–30.
Chen Z, Yin X, Li K, Chen S, Li H, Li Y, Zhang Q, Wang H, Qiu Y. Serum levels of TRIM72 are lower among patients with colon cancer: identification of a potential diagnostic marker. Tohoku J Exp Med. 2018;245(1):61–8.
Kalimuthu S, Zhu L, Oh JM, Lee HW, Gangadaran P, Rajendran RL, Baek SH, Jeon YH, Jeong SY, Lee SW, et al. Regulated mesenchymal stem cells mediated colon cancer therapy assessed by reporter gene based optical imaging. Int J Mol Sci. 2018;19(4):1002.
Zhang R, Men K, Zhang X, Huang R, Tian Y, Zhou B, Yu C, Wang Y, Ji X, Hu Q, et al. Delivery of a modified mRNA encoding IL-22 binding protein (IL-22BP) for colon cancer gene therapy. J Biomed Nanotechnol. 2018;14(7):1239–51.
Mollinari C, Kleman J-P, Jiang W, Schoehn G, Hunter T, Margolis RL. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol. 2002;157(7):1175–86.
Jiang W, Jimenez G, Wells NJ, Hope TJ, Fukunaga R. PRC1: A human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell. 1999;2(6):877–85.
Zhu C, Lau E, Schwarzenbacher R, Bossy-Wetzel E, Jiang W. Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc Natl Acad Sci. 2006;103(16):6196–201.
Kieserman EK, Glotzer M, Wallingford JB. Developmental regulation of central spindle assembly and cytokinesis during vertebrate embryogenesis. Curr Biol. 2008;18(2):116–23.
Brynychova V, Ehrlichova M, Hlavac V, Nemcova-Furstova V, Pecha V, Leva J, Trnkova M, Mrhalova M, Kodet R, Vrana D, et al. Genetic and functional analyses do not explain the association of high PRC1 expression with poor survival of breast carcinoma patients. Biomed Pharmacother. 2016;83:857–64.
Zhang B, Shi X, Xu G, Kang W, Zhang W, Zhang S, Cao Y, Qian L, Zhan P, Yan H, et al. Elevated PRC1 in gastric carcinoma exerts oncogenic function and is targeted by piperlongumine in a p53-dependent manner. J Cell Mol Med. 2017;21(7):1329–41.
Luo HW, Chen QB, Wan YP, Chen GX, Zhuo YJ, Cai ZD, Luo Z, Han ZD, Liang YX, Zhong WD. Protein regulator of cytokinesis 1 overexpression predicts biochemical recurrence in men with prostate cancer. Biomed Pharmacother. 2016a;78(5):116–20.
Yun HJ, Cho YH, Moon Y, Park YW, Yoon HK, Kim YJ, Cho SH, Lee YI, Kang BS, Kim WJ, et al. Transcriptional targeting of gene expression in breast cancer by the promoters of protein regulator of cytokinesis 1 and ribonuclease reductase 2. Exp Mol Med. 2008;40(3):345–53.
Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38(9):1043–8.
Yu W, Feng S, Xing GH, Ping X, Na Z, Yin YF, Sun SY, Jing H, Ying W, Xuan SY. Protein regulator of cytokinesis PRC1 confers chemoresistance and predicts an unfavorable postoperative survival of hepatocellular carcinoma patients. J Cancer. 2017;8(5):801–8.
Schoppmann SF, Vinatzer U, Popitsch N, Mittlböck M, Liebmann-Reindl S, Jomrich G, Streubel B, Birner P. Novel clinically relevant genes in gastrointestinal stromal tumors identified by exome sequencing. Clin Cancer Res. 2013;19(19):5329–39.
Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, Paty PB, Gerald WL, Notterman DA, Domany E. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci USA. 2009;106(17):7131–6.
Stracquadanio G, Wang X, Wallace MD, Grawenda AM, Zhang P, Hewitt J, Zeron-Medina J, Castro-Giner F, Tomlinson IP, Goding CR, et al. The importance of p53 pathway genetics in inherited and somatic cancer genomes. Nat Rev Cancer. 2016;16(4):251–65.
Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8(5):387–98.
Felty Q, Singh KP, Roy D. Estrogen-induced G1/S transition of G0-arrested estrogen-dependent breast cancer cells is regulated by mitochondrial oxidant signaling. Oncogene. 2005;24(31):4883–93.
Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA, Bujnicki JM, Menigatti M, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5(12):1263–75.
Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A “metastasis-prone” signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Cli Exp Metastasis. 2010;27(2):83–90.
Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, Pachlewski J, Oledzki J, Ostrowski J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS ONE. 2010;5(10):e13091.
van Meerloo J, Kaspers GJ, Cloos J. Cell sensitivity assays: the MTT assay. Methods Mol Bio. 2011;731:237–45.
Luo HW, Chen QB, Wan YP, Chen GX, Zhuo YJ, Cai ZD, Luo Z, Han ZD, Liang YX, Zhong WD. Protein regulator of cytokinesis 1 overexpression predicts biochemical recurrence in men with prostate cancer. Biomed Pharmacother. 2016b;78:116–20.
Bu H, Li Y, Jin C, Yu H, Wang X, Chen J, Wang Y, Ma Y, Zhang Y, Kong B. Overexpression of PRC1 indicates a poor prognosis in ovarian cancer. Int J Oncol. 2020;56(3):685–96.
Canman JC, Cabernard C. Mechanics of cell division and cytokinesis. Mol Biol Cell. 2018;29(6):685–6.
Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, Gerlich DW. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136(3):473–84.
Li J, Dallmayer M, Kirchner T, Musa J, Grünewald TGP. PRC1: Linking cytokinesis, chromosomal instability, and cancer evolution. Trends Cancer. 2018;4(1):59–73.
Cho YC, Park J, Park BC, Kim JH, Jeong D, Park S, Cho S. Cell cycle-dependent Cdc25C phosphatase determines cell survival by regulating apoptosis signal-regulating kinase 1. Cell Death Differ. 2015;22(10):1605–17.
Peter M, Peuch C, Labbé J-C, Meyer A, Donoghue D, Doree M. Initial activation of cyclin-B1-cdc2 kinase requires phosphorylation of cyclin B1. EMBO Rep. 2002;3:551–6.
Tane S, Ikenishi A, Okayama H, Iwamoto N, Nakayama K, Takeuchi T. CDK inhibitors, p21(Cip1) and p27(Kip1), participate in cell cycle exit of mammalian cardiomyocytes. Biochem Biophys Res Commun. 2013;443(3):1105–9.
Mortezaee K, Najafi M, Farhood B, Ahmadi A, Potes Y, Shabeeb D, Musa AE. Modulation of apoptosis by melatonin for improving cancer treatment efficiency: an updated review. Life Sci. 2019;228:228–41.
Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182(3):311–22.
Juríková M, Danihel Ľ, Polák Š, Varga I. Ki67, PCNA, and MCM proteins: markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118(5):544–52.
Acknowledgements
None.
Funding
This work was supported by The Natural Science Foundation of Inner Mongolia Autonomous Region [grant number 2019MS08085 and 2018LH08002]; Inner Mongolia People's Hospital Research Fund [grant number 2020YN06]; Start Up Fund for Doctoral Research of People's Hospital of Inner Mongolia Autonomous Region [grant number 2020BS06]; and Inner Mongolia Autonomous Region Health and Family Planning Research Project [grant number 201703011].
Author information
Authors and Affiliations
Contributions
Conception and design of the research: TX, XW; acquisition of data: XJ, FG; analysis and interpretation of data: JL, XJ, FG; statistical analysis: WG, PZ; drafting the manuscript: TX, XW; revision of manuscript for important intellectual content: WJ, PZ. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study obtained permission from the Research Ethics Committee of Inner Mongolia People’s Hospital.
Consent for publication
The patients included in this study had signed informed consents.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
The clinical characteristics of included patients (n = 40). Table S2. Colon cancer clinical and pathological data of patients from tissue microarray (n = 90).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xu, T., Wang, X., Jia, X. et al. Overexpression of protein regulator of cytokinesis 1 facilitates tumor growth and indicates unfavorable prognosis of patients with colon cancer. Cancer Cell Int 20, 528 (2020). https://doi.org/10.1186/s12935-020-01618-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-020-01618-9