Abstract
Background
Lung adenocarcinoma is metastatic cancer with a high mortality rate. Circular RNAs (circRNAs) are a type of noncoding RNA and play a vital role in cancer progression. However, the expression and function of circRNAs in lung adenocarcinoma are still mostly unknown.
Methods
In this study, we screened the differential expression of circRNAs in human bronchial epithelial cells (HBE) and A549 human lung adenocarcinoma cell line (A549) by human circRNA microarray and RT-qPCR. The role of overexpressed circRNA_104889 in A549 cell proliferation, apoptosis, migration, and invasion was studied extensively. Intracellular localization of circRNA_104889 was visualized by FISH assay. MiRNA sponging, ERK1/2 signaling, and caspase-3 expression were analyzed in siRNA-mediated circRNA_104889 knockdowned A549 cells.
Results
CircRNA microarray showed overexpression of circRNA_104889 (> 13-fold) in A459 cells compared to HBE. This finding was further corroborated by the RT-qPCR result. CircRNA_104889 was mainly localized in the cytoplasm of A549 cells. The knockdown of circRNA_104889 in A549 cells by si-RNA mediated RNA interference did not affect cell proliferation and apoptosis but significantly inhibited cell migration and invasion in vitro. Furthermore, knockdown of circRNA_104889 led to an increase of miR4458 expression. Overexpression of miR4458 inhibited A549 cell migration. Both the knockdown of circRNA_104889 and overexpression of miR4458 inhibited the caspase-3 expression and ERK1/2 phosphorylation in A549 cells.
Conclusions
CircRNA_104889 promotes lung adenocarcinoma cell migration and invasion by sponging miR4458 and targeting ERK1/2 signaling and caspase-3 expression.
Similar content being viewed by others
Background
Lung adenocarcinoma is the most common pathological cancer with a high mortality rate [1]. Lung cancer is mainly categorized as small cell and non-small cell lung cancer (NSCLC). Pulmonary adenocarcinoma and pulmonary squamous cell carcinoma are the two main types of NSCLC. Around 85% of lung cancers are NSCLC, and pulmonary adenocarcinoma is the most common NSCLC [2]. Although cancer treatment strategies have been improved, the survival rate of overall lung cancer is only 19% [3]. Multiple factors such as late-stage diagnosis, drug resistance, and metastasis cause poor cancer prognosis and high mortality. Lung adenocarcinoma shows advanced local invasion and distant metastases at the time of diagnosis [4]. Therefore it is crucial to explore the molecular mechanisms of lung adenocarcinoma metastasis to unravel the cost-effective diagnostic and therapeutic markers.
Circular RNAs (circRNAs) is a large family of non-coding RNAs (ncRNAs) circularized by joining the 3′ end of the RNA to the 5′ end, forming a circular structure [5,6,7,8,9]. The lack of accessible ends in circRNAs gives higher resistance to exonucleases compared to linear RNA isoforms [10]. MicroRNAs (miRNAs) are small (20–24 nucleotides) ncRNAs that regulate ~ 30% protein-coding gene in mammals [11]. MiRNAs play a role in post-transcriptional regulation by latching themselves onto mRNAs and inhibiting protein translation by competitive or noncompetitive fashion [12]. MiRNA sponges are competitive inhibitors containing multiple tandem binding sites to the miRNA of interest and effectively obstruct its post-transcriptional regulatory mechanism. CircRNAs had been reported to serve as miRNA ‘sponges’ by combining and encapsulating microRNAs and regulate gene expression [10, 12]. Recent literature have indicated that circRNAs exhibit a powerful functional potential in regulating cancer cell proliferation, apoptosis, migration, and invasion [13,14,15,16], suggesting that circRNAs as a critical regulator in cancer progression. Yao et al. have reported that the close correlation between the circRNA_100876 upregulation and lymph node metastasis and tumor staging of lung adenocarcinoma [17]. Similarly, circ_0026134 has been reported to regulate NSCLC cell proliferation and invasion via sponging miR-1256 and miR-1287 [18]. However, the expression pattern of other circRNAs and their role in lung adenocarcinoma progression and metastasis are still unknown.
MiRNAs regulate a variety of essential biological functions such as cell proliferation, differentiation, apoptosis, and motility. CircRNAs negatively regulate miRNAs function [10, 12]. This ability of circRNAs to compete with the miRNAs could provide the RNA-based targets for cancer therapy. MiR224 expression level negatively correlates with the expression level of caspase-3 in lung cancer tissue samples [19]. Caspase-3 has the regulatory function in colon cancer cell migration, invasion, and metastasis [20]. However, the role of caspase-3 in circRNAs-mediated lung adenocarcinoma progression is still unknown. CircRNAs plays a crucial role in various cancer progressions by regulating the MAPK/ERK1/2 signaling pathway [21]. Therefore, this study focuses on unraveling the possible involvement of circRNA, miRNA, caspase-3, and MAPK/ERK1/2 signaling in adenocarcinoma progression and metastasis.
In the present study, circRNA_104889 showed higher upregulation in lung adenocarcinoma cell line A549 compared to HBE. siRNA-mediated knockdown of circRNA_104889 upregulated miR4458 expression and inhibited A549 cell migration. MiR4458 overexpression inhibited A549 cell invasion and migration, possibly via downregulation of caspase-3 expression and EKR1/2 phosphorylation. The finding of this study proposes a new possible regulatory pathway in lung adenocarcinoma metastasis, i.e., circRNA_104889 regulates lung adenocarcinoma cells metastasis possibly by targeting ERK1/2 signaling and caspase-3 expression via sponging miR4458.
Materials and methods
Cell culture and transfection
HBE and the A549 cells were obtained from Guangzhou Institute of Respiratory Disease. These cell lines were maintained in DMEM/F12 (Invitrogen, CA, US) in the presence of 10% fetal bovine serum, in a humidified 5% CO2 at 37 °C. The miR4458 mimics and negative control were obtained from Invitrogen. The siRNA control (siN05815122147), siRNA1, siRNA2, and siRNA3 obtained from RiboBio Company (Guangzhou, China) are listed in Table 1. MiR4458 mimic, negative control and siRNA were transfected by GenMute™ reagent (Signagen, MD, US) at a final concentration of 50 nM. Total RNA and protein were collected 48 and 72 h after transfection.
CircRNA microarray assay
Arraystar human circRNA array analysis was used to analyze the differential expression pattern of circRNAs in HBE and A549 cells. Total RNA was quantified using the NanoDrop ND-1000 (Thermo Scientific, US). The sample preparation and microarray hybridization were performed based on the Arraystar’s standard protocols. Briefly, total RNA from each sample was amplified and transcribed into fluorescent complementary RNA (cRNA) utilizing random primer according to Arraystar’s Super RNA Labeling protocol (Arraystar Inc, MD, US). The labeled cRNAs were hybridized onto the Arraystar Human circRNA Array (6 × 7 K, Arraystar Inc). The arrays were scanned using the Axon GenePix 4000B microarray scanner (Molecular Devices, MA, US). Scanned images were then imported into Axon Laboratory GenePix Pro 6.0 software for grid alignment and data extraction. Quantile normalization and subsequent data processing were performed using the R software package. Differentially expressed circRNAs with statistical significance between two groups were identified through Volcano Plot filtering. Differentially expressed circRNAs between two samples were identified through fold change filtering. Hierarchical clustering was performed to show the distinct circRNAs expression pattern among samples.
RT-qPCR and RT-PCR analysis
Total RNA from HBE and A459 cells was isolated using the TRIzol® reagent (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized from total RNA using SuperScript III® (Invitrogen) and random primers. Small RNA was extracted by using the RNAiso small RNA kit (Takara, Japan). Reverse transcription reaction was performed using the Mir-X™ miRNA First-Strand Synthesis Kit (Clontech, Japan). The relative circRNA, mRNA, and miRNA levels were quantified with SYBR Premix Ex Taq Kit (Takara) on the CFX96™ Real-Time system (Biorad, US). CircRNA RT-PCR was performed using Bio-rad/T100 and Premix Taq (Takara Taq Version 2.0 plus dye, Japan) according to the manufacturer’s instruction. GAPDH and U6 were used as the internal reference housekeeping gene. The primers used for PCR amplification are listed in Table 2.
Electrophoresis and Sanger sequencing
RT-PCR products were visualized after electrophoresis on 2% agarose gel. The PCR products were purified through the GO-GELU-2500 kit (Geneon BioTech) to confirm the PCR results. Sanger sequencing was performed using Applied Biosystems 3730/3730xl DNA Analyzer (Life Technologies, US). The reverse primer of circRNA_104889 was used for sequencing the product.
Fluorescence in situ hybridization (FISH)
The circRNA_104889 probe: 5′ CATACAGCCCTGTGTGGAGAGTC 3′, and the reverse complement to the circRNA_104889 probe: 5′ GACTCTCCACACAGGGCTGTATG 3′ were used as a junction probe and a negative control probe, respectively. Cells and fluorescein isothiocyanate-labeled RNA probe (Biosense, Guangzhou, China) were denatured at 73 °C for 3 min and incubated at 37 °C for 15–17 h at final concentration 20 ng/μL, then washed (three times for 5 min each) at 43 °C in 50% formamide and 2 × SSC, followed by two 5 min wash in 2 × SSC (the penultimate wash containing DAPI) and a brief washing in nuclease-free water. Finally, a laser scanning confocal microscope (Leica TCS SP8, Germany) was used to take the pictures.
Cell proliferation assay
Cell proliferation assay was conducted using the cell counting kit-8 (CCK-8, Dojindo, Japan). In 96-well, 100 μL of A549 cell suspension (4 × 104 cells/mL) was aliquoted into each well and transfected after 24 h. Then cell proliferation was evaluated at 24, 48, and 72 h after transfection. 10 μL of CCK-8 solution was added to each well, and the plate was incubated in the 37 °C incubator for 2 h, the absorbance of each well was read at 450 nm using a Multiskan™FC (Thermo Scientific).
Migration and invasion assay
For invasion assay, the upper chamber of transwell filter insert with a pore size of 8 μm (Corning Incorporated, NY, US) was coated with 30 mg/cm2 Matrigel (ECM gel, Sigma-Aldrich, St. Louis, MO, US). A total of 4 × 105 cells in 200 μL DMEM/F12 with no FBS were added to the upper chamber. In the lower part of the chamber, 600 μL F12/DMEM with 10% FBS was added, and the assay was performed for 24 h at an atmosphere of 37 °C and 5% CO2. Invasive cells were counted by crystal violet (0.1%) staining, followed by the observation under an inverted microscope, Leica DMI3000B (Leica, Germany). Five random visual fields were counted for each well, and the average was determined. All assays were performed in triplicate and independently repeated three times. The migration assay was performed in the same way with invasion assay, except the matrigel was not used and the number of cells added to the upper chamber was 1 × 105.
Flow cytometry-based apoptosis and cell cycle assays
For cell apoptosis analysis, cells were labeled with Annexin V-FITC and propidium iodide (PI) using an apoptosis detecting kit (BD Biosciences, US) following the manufacturer’s instructions. The stained cells (1 × 105) were then analyzed by a FACSCanto™ II Flow cytometer (BD Biosciences). For cell cycle analysis, the cells were fixed in 70% ice-cold ethanol. The cells were washed with PBS and re-suspended in PI/RNase (1 × 106 cells/0.5 mL) staining buffer. The stained cells were analyzed by flow cytometry.
Luciferase reporter assay
The sequences of circRNA_104889 were synthesized by Invitrogen company and then cloned into the pmiR-RB-REPORT™ by the RiboBio Company (Guangzhou, China). The reporter plasmids (50 nM) and miRNA mimics were co-transfected with Lipofectamine 2000 (Invitrogen) into HEK293 cells (1.5 × 104 cells/well). Cells were collected after 48 h for the analysis using the Dual-Glo®-Luciferase Assay System (Promega, US).
Western blot analysis
Cells were washed with PBS and lysed with RIPA containing protease PMSF and phosphatase inhibitor. The total protein concentration was measured using the BCA protein assay kit (Thermo Scientific). The cell lysate was boiled at 100 °C for 5 min in equal volumes of loading buffer. All samples were subjected to 10% SDS-PAGE, separated by electrophoresis, and transferred to PVDF membranes. After blocking for 2 h in TBST containing 5% non-fat milk, the membranes were incubated overnight at 4 °C with the following primary antibodies: Caspase-3 (#ab13847, Abcam, Cambridge, UK), β-actin(# ab179467, Abcam), ERK (#4695, Cell Signaling Technology, Danvers, MA, USA), Phospho-ERK (#4370, Cell Signaling Technology) diluted in 5% non-fat milk containing TBST. After three times wash, the membranes were incubated with the HRP-conjugated secondary antibodies for 2 h at room temperature. The signals were detected using an enhanced chemiluminescence detection kit (Thermo Scientific, Rockford, IL, USA).
Statistical analysis
Data are presented as the mean ± SD from at least three independent experiments. The differences between the two groups were analyzed with the two-tailed Student’s t-test. Statistical analyses were performed using the SPSS software version 22.0 (IBM Corporation, Armonk, NY, USA), and p < 0.05 was considered statistically significant.
Results
General profile of circRNA microarray
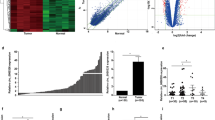
CircRNA microarray test analyzed the differential expressions of circRNA in A549 and HBE cells. The results showed 966 upregulated and 272 downregulated circRNAs in A549 cells compared to HBE. The raw data of differential expression patterns of circRNA in these two cells are listed in Additional files 1, 2. The hierarchical clustering of a portion of circRNA expression in HBE and A549 cells are shown in Fig. 1a. CircRNA_104889 was one of the prominent differentially overexpressed circRNAs in A549 cells compared to HBE. CircRNA_104889 was overexpressed by > 13-fold in A549 cells compared to HBE.
A549 cells showed overexpression of circRNA_104889 with cytoplasmic localization. a The result from hierarchical clustering shows a part of circRNAs expression levels in A5459 cell and HBE cell. b CircRNAs expression by RT-qPCR, data are presented as mean ± SD from 3 independent experiments in triplicates, n = 3. Significant difference between the groups, *p < 0.05. c The nucleic acid electrophoresis result of GAPDH and circRNA_104889 RT-PCR products. d Sanger sequencing of the nucleic acid electrophoresis product by using circRNA_104889 reverse primer. e Analysis of circRNA_104889 distribution in A549 cells by RNA fluorescence in situ hybridization (FISH)
CircRNA_104889 is overexpressed in A549 cells and mainly localizes in the cytoplasm
To validate the results of the circRNA microarray, we selected eight differentially expressed circRNAs associated with cancer. The association of these circRNAs with cancer was predicted by bioinformatics (Targetscan, miRDB [22], and starBase v2.0 [23]). RT-qPCR was performed to confirm eight circRNAs expression levels in HBE and A549 cells. The data showed that circRNA_101099 was downregulated while the circRNA_102633, circRNA_101972, and circRNA_104889 were upregulated in A549 compared to HBE. The RT-qPCR result showed a 22-fold higher expression of circRNA_104889 in A549 cells compared to HBE (Fig. 1b). Head-to-tail splicing of circRNA_104889 was assayed and validated by nucleic acid electrophoresis and Sanger sequencing (Fig. 1c, d). Then circRNA_104889 reverse primer was provided for sequencing the product, and the sequences showed a definite crossing of the head-to-tail splicing site (Fig. 1d). CircRNA_104889 sequencing according to the human reference genome (GRCh37/hg19) obtained from the UCSC genome database (http://genome.ucsc.edu/) showed 1063 bp genomic length and spliced length of the circRNA-104889. CircRNA-104889 is located at chr9(+):118949432–118950495, and its associated gene symbol is PAPPA (Pregnancy-associated plasma protein A) exon 2.
To evaluate the subcellular localization of circRNA_104889, RNA-fluorescence in situ hybridization (FISH) was performed in A549 cells. The results revealed that circRNA_104889 was mainly distributed in the cytoplasm (Fig. 1e), suggesting its possible role in post-transcriptional regulation.
CircRNA_104889 promotes the migration and invasion of lung adenocarcinoma cells
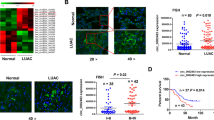
CircRNA_104889 was knockdowned in A549 cells using siRNA-mediated gene silencing (Fig. 2a). RT-qPCR confirmed that siRNA2 inhibits circRNA_104889 expression by 74.3% compared to the scrambled control (Fig. 2b). Furthermore, a 24 h transfection of siRNA2 showed the highest circRNA_104889 knockdown potential (Fig. 2c). The siRNA2 downregulates the expression of circRNA_104889 and could decrease its parental gene pregnancy-associated plasma protein A (PAPPA) (Fig. 2d).
Downregulation of circRNA_104889 in vitro. a Three siRNAs targeting the circRNA_104889 in junction sequences (the red arrows point to the head-to-tail splicing site) were designed. b The expression level of circRNA_104889 in A549 cells after transfection with three siRNAs. c Expression of circRNA_104889 after siRNA2 transfection for 24, 48, and 72 h in A549 cells. d The mRNA levels of the parental gene PAPPA after knockdown of circRNA_104889 with siRNA2. Data are presented as mean ± SD from 3 independent experiments in triplicates, n = 3. Significant effect of the treatments, *p < 0.05, **p < 0.01, ***p < 0.001
To validate the role of circRNA_104889 dysregulation in the tumorigenesis and metastasis of lung adenocarcinoma, we analyzed cell proliferation, migration, invasion, apoptosis, and cell cycle in A559 cells with or without circ_104889 knockdown. The proliferation of circRNA_104889 knockdowned cells was slightly decreased (Fig. 3a). Besides, the knockdown of circRNA_104889 increased the percentage of G2/M cells (Fig. 3b) and apoptotic rate (Fig. 3c). However, the difference was not statistically significant. An evident decrease in migration (Fig. 3d) and invasion (Fig. 3e) were observed in circRNA_104889 knockdowned A549 cells (p < 0.05) compared to the control group. These results suggest the possible role of circRNA_104889 in lung cancer metastasis.
CircRNA_104889 competes with miR4458 and targets caspase-3
Target prediction in miRDB, starBase v2.0, and TargetScan showed that the miR4458 has two binding sites in circRNA_104889. The 3′ UTR of the caspase-3 gene contains separate miR4458 binding seed sequences that are conserved through evolution (Fig. 4a). RT-qPCR data showed the overexpression of miR4458 in circRNA_104889 knockdowded A549 (Fig. 4b). To validate whether circRNA_104889 is a bona fide target for miR4458 or not, we inserted the whole sequence of circRNA_104889 into the 3′ UTR locus of the firefly luciferase gene. It demonstrated that miR-4458 dramatically suppressed the luciferase activity in HEK293 cells (Fig. 4c). Subsequently, ectopic expression of miR4458 in A549 cells transfected with miR-4458 mimics also led to a significant reduction in cell invasion (Fig. 4d). We further detected the mRNA level of caspase-3 in circRNA_104889 knockdowned (Fig. 5a) and miR4458 overexpressed A549 cells (Fig. 5b). The caspase-3 mRNA expression was downregulated in both conditions (Fig. 5a, b). The protein level of caspase-3 was subsequently detected by western blot (Fig. 5c, d). Both the knockdown of circRNA_104889 and the overexpression of miR4458 in A549 cells inhibited caspase-3 expression. The results were consistent with the changes in the mRNA level of caspase-3 (Fig. 5a–d).
Interaction between circRNA_104889 and miR4458. a Schematic representation of two miRNA binding sites (labeled blue arc and red base) in circRNA_104889, while the 3′UTR region of caspase-3 harbors a putative miR4458 binding site, which is highly conserved across multiple species. b The change of miR4458 expression after downregulating circRNA_104889 with siRNA2. c Dual-luciferase reporter assays using reporter constructs with the entire circRNA_104889 sequence, then HEK293 cells were co-transfected with pmiR-RB-REPORT™ (WT) and either miR4458 or control mimics. d Cell invasion assay. Data are presented as mean ± SD from 3 independent experiments in triplicates, n = 3. Significant effect of the treatments, ** p < 0.01, *** p < 0.001
CircRNA_104889 activates the ERK1/2 pathway by up-regulating the caspase-3. a, b The mRNA level of the caspase-3 after knockdown of circRNA_104889 or overexpression of miR4458. c Representative images of western blots of caspase-3. d Quantification of caspase-3 protein expression from western blots. e Representative images of western blots of ERK1/2 and p-ERK1/2. f Quantification of pERK/ERK from western blots. g The schematic diagram showing the possible molecular mechanism of circRNA_104889-mediated metastasis of lung adenocarcinoma. Data are presented as mean ± SD from 3 independent experiments in triplicates, n = 3. Significant effect of the treatments,*p < 0.05, **p < 0.01, ***p < 0.001
CircRNA_104889 activates ERK1/2 pathway
The knockdown of circRNA_104889 significantly reduced the level of phosphorylated ERK1/2, with no detectable changes in expression of total ERK1/2 (Fig. 5e, f). These results suggest circRNA_104889 as an inducer of lung adenocarcinoma metastasis by activating the ERK1/2 pathway.
Discussion
Although non-coding RNAs were initially argued to be spurious transcriptional noise, recent pieces of evidence suggest their important role in cellular development and human diseases [24,25,26]. Circ-ITCH has been reported to reduce cell viability and arrest proliferation in esophageal squamous cell carcinoma [14]. Circ_RNA100783 has been optimized as an immunosenescence biomarker of T-cell aging [26]. Hsa-circ-000595 regulates the apoptosis of aortic smooth muscle cells associated with aortic aneurysm [27]. Additionally, circRNA-CER regulates MMP13 expression and participate in the process of chondrocyte ECM degradation [28]. However, the expression pattern of circRNAs in lung adenocarcinoma and their role in lung cancer progression and metastasis are still unknown. In this study, we revealed the higher expression of circRNA_104889 in lung adenocarcinoma A549 cells. The knockdown of circRNA_104889 inhibited A549 cell migration and invasion. Interestingly, knockdown of circRNA_104889 upregulated miR4458 expression, downregulated caspase-3 expression, and inhibited ERK1/2 phosphorylation. Our findings suggest the possible role of circRNA_104889 in lung adenocarcinoma metastasis by sponging miR4458 expression and possibly via modulation of caspase-3 expression and ERK1/2 phosphorylation (Fig. 5g).
CircRNA_104889 locates at chromosome 9: 118949432–118950495 on the plus strand, aligned in sense orientation to the known protein-coding gene PAPPA and contains only one exon. The length of the circRNA_104889 is 1063 bp. Jeck et al. have reported that circRNAs tend to accumulate in the cytoplasm of some cells, and may even be expressed higher than the associated linear mRNAs [29]. In this study, the FISH assay showed that circRNA_104889 mainly exists in the cytoplasm of A549 cells, suggesting the possible role of circRNA_104889 in the post-transcriptional regulation. CircRNAs bind to miRNAs, acting as miRNA sponges and regulate gene expression at the transcriptional or post-transcriptional level [12]. It has been reported that hsa_circ_0001855 and hsa_circ_0004904 act as sponges of their miRNA recognition elements (MREs) and block the inhibition of MREs on their target mRNA of the PAPPA gene in preeclampsia patients [30]. TargetScan and miRDB software-based miRNA binding site prediction showed the 3′ UTR region of the PAPPA gene as a binding site for miR4458 within circRNA_104889, indicating that the linear PAPPA positively correlates with levels of circRNA_104889. This sponging of miR445 by circ_104889 might downregulate PAPPA in lung adenocarcinoma.
Extracellular signal-regulated kinase 1/2 (ERK1/2) pathway plays a critical role in tumor development and contributes to cell metastasis [31]. Furthermore, the ERK1/2 pathway is an important signal transduction pathway involving in various cellular activities and diseases [32,33,34,35,36,37,38]. MiR7 has been reported to inhibit ERK1/2 signaling and epithelial growth factor receptor expression in lung cancer, breast cancer, and glioblastoma cell lines [39]. In this study, knockdown of circRNA_104889 in A549 cells inhibited EKR1/2 phosphorylation, upregulated mi4458, and inhibited caspase-3 activity. Moreover, mi4458 mimics inhibited A549 cell migration. MiR4458, a new tumor-suppressor, is downregulated in human hepatocellular carcinoma [40], and colon cancer [41]. Caspase-3 is known as a cysteine protease that primarily executes cell death and also regulates cancer cell migration [19, 20, 42,43,44,45]. Here, we predict that circRNA_104889 and caspase-3 are the common intracellular targets of miR4458. The knockdown of circRNA_104889 in A549 cells significantly increased miR4458 expression and reduced caspase-3 expression. Moreover, it has been reported that the depletion of caspase-3 leads to the inactivation of ERK1/2 signaling and reduces the cell motility and invasion of breast cancer cell line MCF7 [46]. Our findings indicate that circRNA_104889 enhances A549 cell migration by sponging miR4458 possibly, via the upregulation of caspase-3 activity and ERK1/2 phosphorylation.
Salmena et al. suggested the possible role of pseudogene transcripts and long noncoding RNAs (lncRNAs) or other molecules that regulate each other’s expression by using MREs to compete for the binding of microRNAs and affect the cell functions [47]. Pseudogene transcripts, lncRNAs, mRNAs, and circRNAs are reported to be involved in tumorigenesis and development of human cancer via competing endogenous RNA (ceRNA) mechanism [48,49,50,51,52,53]. The involvement of the ceRNA mechanism in circRNA_104889 mediated sponging of mi4458 should be further investigated. A limitation of this study is that although the knockdown of circRNA_104889 inactivated the ERK1/2 signaling via inhibiting caspase-3 activity, the exact role of caspase-3 and ERK1/2 signaling in A549 migration and invasion have not been thoroughly investigated. Further studies are recommended to unravel the role of caspase-3 and ERK1/2 in lung adenocarcinoma metastasis using lung adenocarcinoma tissue samples, lung cancer cell lines, and patient-derived xenograft model of lung adenocarcinoma. Another limitation of this study is the lack of in vivo studies elucidating the role of circRNA_104889 in lung adenocarcinoma invasion and metastasis. Validation of the results of this study in the patient-derived xenograft mice model of lung adenocarcinoma is strongly recommended.
Conclusions
We found the upregulation of circRNA_104889 in lung adenocarcinoma cell line A549. The knockdown of circRNA_104889 inhibited A549 cell migration and invasion by sponging miR4458. Moreover, knockdown of circRNA_104889 inhibited the caspase-3 expression and ERK1/2 phosphorylation via miR4458 sponging. Our findings implie circRNA_104889 is an oncogenic circRNA that promotes migration and invasion of lung adenocarcinoma cells.
Availability of data and materials
The data are included within the manuscript.
Abbreviations
- CircRNAs:
-
Circular RNA
- HBE:
-
Human bronchial epithelial cells
- A549:
-
A549 human lung adenocarcinoma cell line
- siRNA:
-
Small interfering RNA
- miRNA:
-
MicroRNA
- CKD2:
-
Cycle proteins cyclin-dependent kinase 2
- p21:
-
Cyclin-dependent kinase inhibitor 1
- FISH:
-
Fluorescence in situ hybridization
- PI:
-
Propidium iodide
- lncRNA:
-
Long non-coding RNA
- ceRNA:
-
Competing endogenous RNA
- cRNA:
-
Complementary RNA
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–46.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.
Williams CD, Gajra A, Ganti AK, Kelley MJ. Use and impact of adjuvant chemotherapy in patients with resected non-small cell lung cancer. Cancer Am Cancer Soc. 2014;120(13):1939–47.
AbouHaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci USA. 2014;111(40):14542–7.
Haimovich G, Medina DA, Causse SZ, Garber M, Millan-Zambrano G, Barkai O, et al. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153(5):1000–11.
Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–7.
Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–61.
Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340(6131):440–1.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8.
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8.
Rong DW, Tang WW, Li ZX, Zhou J, Shi JF, Wang HJ, et al. Novel insights into circular RNAs in clinical application of carcinomas. Oncotargets Ther. 2017;10:2183–8.
Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–65.
Han JQ, Zhao GB, Ma X, Dong Q, Zhang H, Wang Y, et al. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Co. 2018;503(4):2429–35.
Fan HQ, Li YY, Liu CD, Liu YM, Bai J, Li W. Circular RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem Biophys Res Co. 2018;507(1–4):178–84.
Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–6.
Chang H, Qu J, Wang J, Liang X, Sun W. Circular RNA circ_0026134 regulates non-small cell lung cancer cell proliferation and invasion via sponging miR-1256 and miR-1287. Biomed Pharmacother. 2019;112:108743.
Cui R, Kim T, Fassan M, Meng W, Sun HL, Jeon YJ, et al. MicroRNA-224 is implicated in lung cancer pathogenesis through targeting caspase-3 and caspase-7. Oncotarget. 2015;6(26):21802–15.
Zhou M, Liu XJ, Li ZH, Huang Q, Li F, Li CY. Caspase-3 regulates the migration, invasion and metastasis of colon cancer cells. Int J Cancer. 2018;143(4):921–30.
Yang ZZ, Xie L, Han L, Qu X, Yang YH, Zhang Y, et al. Circular RNAs: regulators of cancer-related signaling pathways and potential diagnostic biomarkers for human cancers. Theranostics. 2017;7(12):3106–17.
Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database issue):D146–52.
Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7.
Irizar H, Munoz-Culla M, Saenz-Cuesta M, Osorio-Querejeta I, Sepulveda L, Castillo-Trivino T, et al. Identification of ncRNAs as potential therapeutic targets in multiple sclerosis through differential ncRNA–mRNA network analysis. BMC Genomics. 2015;16:250.
Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–88.
Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing. 2015;12:17.
Zheng C, Niu H, Li M, Zhang H, Yang Z, Tian L, et al. Cyclic RNA hsacirc000595 regulates apoptosis of aortic smooth muscle cells. Mol Med Rep. 2015;12(5):6656–62.
Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, et al. Circular RNA related to the Chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘sponge’ in human cartilage degradation. Sci Rep. 2016;6:22572.
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu JZ, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–57.
Jiang M, Lash GE, Zhao XQ, Long Y, Guo CJ, Yang HL. CircRNA-0004904, CircRNA-0001855, and PAPP-A: potential novel biomarkers for the prediction of preeclampsia. Cell Physiol Biochem. 2018;46(6):2576–86.
Xander P, Brito RR, Perez EC, Pozzibon JM, de Souza CF, Pellegrino R, et al. Crosstalk between B16 melanoma cells and B-1 lymphocytes induces global changes in tumor cell gene expression. Immunobiology. 2013;218(10):1293–303.
Ai X, Wu Y, Zhang W, Zhang Z, Jin G, Zhao J, et al. Targeting the ERK pathway reduces liver metastasis of Smad4-inactivated colorectal cancer. Cancer Biol Ther. 2013;14(11):1059–67.
Gao J, Liu X, Yang F, Liu T, Yan Q, Yang X. By inhibiting Ras/Raf/ERK and MMP-9, knockdown of EpCAM inhibits breast cancer cell growth and metastasis. Oncotarget. 2015;6(29):27187–98.
Ha GH, Park JS, Breuer EK. TACC3 promotes epithelial-mesenchymal transition (EMT) through the activation of PI3K/Akt and ERK signaling pathways. Cancer Lett. 2013;332(1):63–73.
Miao JH, Wang SQ, Zhang MH, Yu FB, Zhang L, Yu ZX, et al. Knockdown of galectin-1 suppresses the growth and invasion of osteosarcoma cells through inhibition of the MAPK/ERK pathway. Oncol Rep. 2014;32(4):1497–504.
Wu S, Lao XY, Sun TT, Ren LL, Kong X, Wang JL, et al. Knockdown of ZFX inhibits gastric cancer cell growth in vitro and in vivo via downregulating the ERK-MAPK pathway. Cancer Lett. 2013;337(2):293–300.
Yang D, Wang LL, Dong TT, Shen YH, Guo XS, Liu CY, et al. Progranulin promotes colorectal cancer proliferation and angiogenesis through TNFR2/Akt and ERK signaling pathways. Am J Cancer Res. 2015;5(10):3085–97.
Zeng Z, Leng T, Feng X, Sun H, Inoue K, Zhu L, et al. Silencing TRPM7 in mouse cortical astrocytes impairs cell proliferation and migration via ERK and JNK signaling pathways. PLoS ONE. 2015;10(3):e0119912.
Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284(9):5731–41.
Tang D, Sun B, Yu H, Yang Z, Zhu L. Tumor-suppressing effect of miR-4458 on human hepatocellular carcinoma. Cell Physiol Biochem. 2015;35(5):1797–807.
Qin Y, Cheng C, Lu H, Wang Y. miR-4458 suppresses glycolysis and lactate production by directly targeting hexokinase2 in colon cancer cells. Biochem Biophys Res Commun. 2016;469(1):37–43.
Ding D, Qi W, Yu D, Jiang H, Han C, Kim MJ, et al. Addition of exogenous NAD + prevents mefloquine-induced neuroaxonal and hair cell degeneration through reduction of caspase-3-mediated apoptosis in cochlear organotypic cultures. PLoS ONE. 2013;8(11):e79817.
Mai L, Yi F, Gou X, Zhang J, Wang C, Liu G, et al. The overexpression of MCPH1 inhibits cell growth through regulating cell cycle-related proteins and activating cytochrome c-caspase 3 signaling in cervical cancer. Mol Cell Biochem. 2014;392(1–2):95–107.
Tourigny A, Charbonneau F, Xing P, Boukrab R, Rousseau G, St-Arnaud R, et al. CYP24A1 exacerbated activity during diabetes contributes to kidney tubular apoptosis via caspase-3 increased expression and activation. PLoS ONE. 2012;7(10):e48652.
Yu Z, Zhang C, Wang H, Xing J, Gong H, Yu E, et al. Multidrug resistance-associated protein 3 confers resistance to chemoradiotherapy for rectal cancer by regulating reactive oxygen species and caspase-3-dependent apoptotic pathway. Cancer Lett. 2014;353(2):182–93.
Cheng YJ, Lee CH, Lin YP, Huang JY, Su CC, Chang WT, et al. Caspase-3 enhances lung metastasis and cell migration in a protease-independent mechanism through the ERK pathway. Int J Cancer. 2008;123(6):1278–85.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–8.
Fang L, Du WW, Yang X, Chen K, Ghanekar A, Levy G, et al. Versican 3′-untranslated region (3′-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB J. 2013;27(3):907–19.
Lee DY, Jeyapalan Z, Fang L, Yang J, Zhang Y, Yee AY, et al. Expression of versican 3′-untranslated region modulates endogenous microRNA functions. PLoS ONE. 2010;5(10):e13599.
Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147(2):370–81.
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–57.
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–83.
Wang L, Guo ZY, Zhang R, Xin B, Chen R, Zhao J, et al. Pseudogene OCT4-pg4 functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 in hepatocellular carcinoma. Carcinogenesis. 2013;34(8):1773–81.
Acknowledgements
Not applicable.
Funding
This work was supported by Project of Education of Guangdong Province (2017KQNCX162), Project of Guangzhou Municipal Health Commission (20181A011103) and the Project of Liwan District Science and Technology (201804015).
Author information
Authors and Affiliations
Contributions
YY, JY and JLP contributed equally to this study and shared the first authorship. YY, JY, JLP: experiment, data analysis and manuscript preparation; HW, JZ, YW: data analysis; HT, LG: Study design, data interpretation, manuscript revision; All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for Publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Differentially upregulated circRNAs in A549 cells compared to HBE.
Additional file 2.
Differentially downregulated circRNAs in A549 cells compared to HBE.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yan, Y., Yang, J., Pathak, J.L. et al. CircRNA_104889 promotes lung adenocarcinoma cell invasion via sponging miR4458. Cancer Cell Int 20, 432 (2020). https://doi.org/10.1186/s12935-020-01522-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-020-01522-2