Abstract
Background
Pulmonary malignant neoplasms have a high worldwide morbidity and mortality, so the study of these malignancies using microRNAs (miRNAs) has attracted great interest and enthusiasm. The aim of this study was to determine the clinical effect of hsa-microRNA-204-5p (miR-204-5p) and its underlying molecular mechanisms in non-small cell lung cancer (NSCLC).
Methods
Expression of miR-204-5p was investigated by real-time quantitative PCR (RT-qPCR). After data mining from public online repositories, several integrative assessment methods, including receiver operating characteristic (ROC) curves, hazard ratios (HR) with 95% confidence intervals (95% CI), and comprehensive meta-analyses, were conducted to explore the expression and clinical utility of miR-204-5p. The potential objects regulated and controlled by miR-204-5p in the course of NSCLC were identified by estimated target prediction and analysis. The regulatory network of miR-204-5p, with its target genes and transcription factors (TFs), was structured from database evidence and literature references.
Results
The expression of miR-204-5p was downregulated in NSCLC, and the downtrend was related to gender, histological type, vascular invasion, tumor size, clinicopathologic grade and lymph node metastasis (P<0.05). MiR-204-5p was useful in prognosis, but was deemed unsuitable at present as an auxiliary diagnostic or prognostic risk factor for NSCLC due to the lack of statistical significance in meta-analyses and absence of large-scale investigations. Gene enrichment and annotation analyses identified miR-204-5p candidate targets that took part in various genetic activities and biological functions. The predicted TFs, like MAX, MYC, and RUNX1, interfered in regulatory networks involving miR-204-5p and its predicted hub genes, though a modulatory loop or axis of the miRNA-TF-gene that was out of range with shortage in database prediction, experimental proof and literature confirmation.
Conclusions
The frequently observed decrease in miR-204-5p was helpful for NSCLC diagnosis. The estimated target genes and TFs contributed to the anti-oncogene effects of miR-204-5p.
Similar content being viewed by others
Background
The worldwide morbidity and mortality of pulmonary cancer has remained high for decades in both genders, reflecting an increase in contributory factors like tobacco use and air pollution [1,2,3,4,5]. The two primary categories of pulmonary neoplasms are small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), with NSCLC accounting for approximately 80% of all pulmonary cancers. NSCLC includes adenocarcinoma, squamous cell lung carcinoma, undifferentiated large cell carcinoma, adenosquamous carcinoma and bronchioalveolar carcinoma; the first three are the best known types [6]. The survival of patients with NSCLC is still bleak due to delayed diagnosis, undisciplined treatment, incident chemoresistance, and frequent tumor recurrence [7,8,9]. Thus, thorough investigation of the molecular mechanisms underlying lung carcinogenesis remains an urgent task, for establishing new and effective guidelines for cancer screening and for identifying novel genetic targets for treatments.
One potential class of molecular targets are the microRNAs (miRNAs). These are small non-coding RNA molecules, with approximately 20 nucleotides in length, that negatively modulate expression of target genes by completely or incompletely binding to the 3′ untranslated region (UTR) of messenger RNAs (mRNAs) [10,11,12,13]. The miRNAs have been proposed as novel diagnostic biomarkers and prognostic indicators for tumorigenic processes, as they play indispensable roles in cancer cell differentiation, proliferation, and apoptosis, and in metastasis and recurrence of numerous malignant tumors [10, 14]. One miRNA, hsa-microRNA-204-5p (also known as miR-204-5p, or miR-204), has attracted attention in NSCLC research, because its low expression in NSCLC tumors is associated with advanced progression, poor prognosis and severe metastatic potential [15,16,17].
Previous studies on the mechanisms of miR-204-5p on NSCLC has mainly focused on the repression of specific mRNAs, so knowledge about its multilateral functions or its clinical prospects remains limited. Aberrant expression of miR-204-5p is now a well-established feature of pulmonary carcinogenesis; however, what is still unclear is the clinical contribution of miR-204-5p and particularly its potential role in the early detection of NSCLC. The mechanism by which miR-204-5p mediates its target mRNA-protein signaling networks to regulate tumor progression is also not yet established.
The current work describes distinctive features of miR-204-5p expression in NSCLC by integrative analysis of results from real-time quantitative polymerase chain reaction (RT-qPCR) and from sequence and genechip data from the cancer genome atlas (TCGA), Gene Expression Omnibus (GEO), and the current literature, in addition to relevant prediction materials from online tools. Our goals were to explore the possibility that miR-204-5p might be a promising indicator for NSCLC process and to identify our perspective on other underlying regulatory mechanisms at the molecular level (Fig. 1).
Study structure and major research methods in this paper. Abbreviation: GEO, Gene Expression Omnibus; TCGA, the cancer genome atlas; RT-qPCR, real-time quantitative polymerase chain reaction; ROC, receiver operating characteristics; HR, hazard ratio; 95% CI, 95% confidence interval; SMD, standard mean deviation; SROC, summarized receiver operating characteristics; DEGs, differentially expressed genes; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; TF, transcription factor
Methods
Patients and samples
Formalin-fixed, paraffin-embedded (FFPE) samples and corresponding non-cancerous lung tissues were obtained with prior informed consent from125 patients with NSCLC treated at Department of Pathology, the First Affiliated Hospital of Guangxi Medical University (Nanning, Guangxi, China) from January 2012 to February 2014. The research proposal was approved by the Committee on Ethics of the First Affiliated Hospital of Guangxi Medical University. All cases were pathologically distinguished and verified by two recognized experts (Zhen-bo Feng and Gang Chen). Each participant was classified based on pathological pattern, tumor size, and clinicopathologic grade according to the IASLC 2009 criteria [18].
RNA isolation and RT-qPCR
Total RNA was extracted from FFPE samples from the NSCLC and matching tissues by miRNeasy Kit (QIAGEN, KJVenlo, The Netherlands) according to the manual instructions. The RNA concentration was quantified using a NanoDrop 2000 instrument (Wilmington, DE, USA). Then, reverse transcription synthesis of complimentary DNA (cDNA) was conducted on First Strand cDNA Synthesis Kit (Thermo Scientific, USA), followed by PCR reaction on an Applied Biosystems PCR7900 instrument (Thermo Fisher Scientific, Waltham, USA). The thermal cycling steps started at 95 °C for 10 min, continued with totally 40 PCR cycles of 15 s at 95 °C and 60s at 60 °C, finally annealed at 72 °C for 5 s. RNU6B was utilized as the housekeeping miRNA for miR-204-5p. The primer sequences used in the TaqMan® MicroRNA Assays were as follows: RNU6B (Applied Biosystems,4,427,975–001093)-CGCAAGGAUGACACGCAAAUUCGUGAAGCGUUCCAUAUUUUU and miR-204-5p (Applied Biosystems, 4,427,975–000508)- UUCCCUUUGUCAUCCUAUGCCU. The RT-qPCR process was performed on an Applied Biosystems PCR7900 instrument using the protocol supplied by the manufacturer. The expression levels of the two miRNAs were compared using the 2 − ΔΔCt method [19]. All specimens were analyzed in triplicate.
Data mining from TCGA
The Illumina HiSeq miRNA-sequencing data for miR-204-5p were downloaded and extracted from TCGA up to October 31, 2018. The Xena Public Data Hubs online analysis program (https://xena.ucsc.edu/public-hubs/) was used to calculate expression level of miR-204-5p and to assess the difference between 999 NSCLC and 91 normal tissues. The genes involved in NSCLC were also obtained from TCGA data and further analyzed with the EdgeR package. Genes with a false discovery rate (FDR)<0.05 were deemed differentially expressed genes (DEGs) and selected as standby members.
Collection and management of miR-204-5p data
Genechips data related to miR-204-5p in NSCLC were sought in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) up to October 31,2018.To evaluate the clinical application of miR-204-5p for NSCLC, data on documented expression of miR-204-5p between NSCLC and non-tumorous controls were collected from the following databases: PubMed, Web of Science, Wiley online library, Springerlink, Embase, Chinese National Knowledge Infrastructure, Chinese Biomedical Database, Chinese VIP and Wan Fang data resources. The data retrieval entry was as follows: (MicroRNA OR miRNA OR “Micro RNA” OR “Small Temporal RNA” OR “non-coding RNA” OR ncRNA OR “small RNA”) AND (lung OR pulmonary OR respiratory OR bronchi OR bronchioles OR alveoli OR pneumocytes OR “air way”) AND (cancer OR carcinoma OR tumor OR neoplas* OR malignan* OR adenocarcinoma).
The microarray chip data and publications had to fulfill the following conditions for inclusion in the current study:1) the research object focused on human beings, 2) the publication was in English or Chinese, 3) participants were confirmed cases with NSCLC, 4) cases included normal controls and contained at least 2 samples, and 5) pertinent expression of miR-204-5p in NSCLC and corresponding non-tumorous specimens was explored. Exclusion criteria included:1) duplicate selections of studies, conference abstracts, expert opinions, case reports, comments, letters, editorial or reviews, 2) articles with in vitro or in vivo experiments or human xenografts, 3) data with no information about miR-204-5p expression, and 4) publications not written in English or Chinese.
Items from the eligible datasets and reports included for further investigation were: series accession, the lead author, publication year, nationality, experimental platform, sample size, types of sample, research techniques, amount of miR-204-5p and threshold value. The above screening procedures were repeated by two veteran researchers.
Prediction and analyses of miR-204-5p target genes
MiRwalk 2.0, an online miRNA-target search tool that integrates 12 prediction programs (miRWalk, miRanda, miRDB, MicroT4, miRMap, miRNAMap, miRBridge, PITA, PICTAR2, RNAhybrid, RNA22 and TargetScan), was applied to predict the target genes for subsequent analyses. Only genes that co-occurred in at least six databases were deemed eligible. Due to the decrease of miR-204-5p in NSCLC, the target genes were expected to be expressed at a higher level to a large extent, so up-regulated DEGs from TCGA were adopted for further work. The final estimated objects for miR-204-5p were derived from the intersection of online databases and TCGA.
The selected candidate DEGs were then processed in the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8 (https://david-d.ncifcrf.gov/) to obtain the gene ontology (GO) annotation as well as the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. P < 0.05 was regarded as the cut-off. Further information about the interaction between the proteins encoded by DEGs was obtained using the Search Tool for the Retrieval of Interacting Genes (STRING) (http://www.string-db.org/) and Cytoscape 3.6.1 to establish a protein-protein interaction (PPI) network for DEGs that participated in the top three GO items and KEGG pathways. In this study, the selection criterion for hub genes was based on the degree of connection among pitch points in the PPI network. The mRNA expression levels of the hub genes were also accessed from GEPIA (http://gepia.cancer-pku.cn), and their protein variations were validated in the Human Protein Atlas (THPA) (https://www.proteinatlas.org/).
Transcription factor prediction
Transcription factors (TFs) that were likely to related to miRNA-204-5p and/or hub genes were predicted from public databases, followed by collection of experimentally confirmed targets from literature. Relevant TFs that were able to influence miR-204-5p were mainly predicted using three different online databases that provided estimated relationships between TFs and marker genes: Gene Transcription Regulation Database (GTRD, http://gtrd.biouml.org/), HTFtarget database (http://bioinfo.life.hust.edu.cn/hTFtarget#!/) and TransmiR v2.0 database (http://www.cuilab.cn/transmir). The TFs that modulated hub genes were acquired from GTRD and HTFtarget simultaneously. Precise information was obtained from the intersection of the predictions for combinatorial utilization. The relationships between these can be described as TF-miRNA (GTRD ∩ HTFtarget ∩ TransmiR) ∩ TF-hub genes (GTRD ∩HTFtarget). The predicted transcription factor binding sites (TFBSs) were retrieved from the JASPAR database (http://jaspar.genereg.net/), and the sequences were derived from the positive-sense strand with the highest score. Literature mining was performed with combined keywords (MicroRNA OR miRNA OR “Micro RNA” OR “Small Temporal RNA” OR “non-coding RNA” OR ncRNA OR “small RNA”) AND (transcription factor OR transcriptional factor) AND (cancer OR carcinoma OR tumor OR neoplasm* OR malignant* OR adenocarcinoma) to confirm the relationships between motifs and NSCLC or other types of cancers. Synergistic co-regulatory motifs of miR-204-5p network were constructed based on the expected regulation and literature confirmation.
Statistical analysis
Results of miR-204-5p expression were reported as mean ± standard deviation (SD). Student’s t-test was used to compare differences in miR-204-5p expression measured by RT-qPCR or raw expression data. One-way analysis of variance (ANOVA) was conducted to evaluate the characteristics of miR-204-5p distribution among groups including three or more variates. Statistical analyses were performed using SPSS v22.0 (SPSS Inc., Chicago, IL, USA).
Data from GEO were first individually processed for acquisition of standard mean deviation (SMD) by meta-analysis, followed by their integration with TCGA and the literature to evaluate distinct expression and potential application prospects of miR-204-5p. The analytical methods in meta-analyses were identical to those used in previous studies [20, 21], and analysis was conducted using by Stata 12.0 (Stata Corp LP, College Station, USA). The role of miR-204-5p in NSCLC diagnosis was studied using receiver operating characteristic (ROC) and summarized receiver operating characteristics (SROC) curves were respectively constructed in accordance with the previous studies [22].
The RT-qPCR data were divided into low-level and high-level groups in according to the median expression level of miR-204-5p(Median = 3.75). The association between survival data in the two groups and miR-204-5p expression were analyzed by Kaplan-Meier (K-M) curves and univariate Cox regression analysis using SPSS v22.0. The hazard ratio (HR), 95% confidence interval (95% CI), and other data available from public resources were either extracted directly or obtained indirectly by recommendations of Tierney et al. [23]. Comprehensive meta-analysis was then preformed for the HRs to appraise the efficiency of miR-204-5p in NSCLC prognosis.
In this work, P < 0.05 was considered as statistically significant. A random effects model was considered valid when a large heterogeneity was defined with reference to I2 greater than 50% or P less than 0.1; otherwise a fixed- coefficient model was in usage [24,25,26,27].
Results
Differential expression and clinical characteristics of miR-204-5p in NSCLC
Relative quantitative expression and the fundamental characteristics of miR-204-5p in the research subjects are listed in Table 1. The RT-qPCR results (Table 1 and Fig. 2a) showed a statistically significant difference in the quantitative variation of miR-204-5p between NSCLC and normal adjacent tissues (P = 0.001). Statistical differences were also found for gender, tumor size, histological type, vascular invasion, tumor node metastasis (TNM) grade, and lymph node metastasis (P<0.05). Differences of miR-204-5p in expression between pathological types were assessed by analyzing RT-qPCR data grouped into lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) (Table 2, Table 3 and Fig. 2b). Apart from age and smoking behavior, lower of miR-204-5p expression was noted in sex, tumor size, vascular invasion, TNM grade, lymph node metastasis, and pathological grading in the LUAD group than in the LUSC group (P<0.05). Therefore, miR-204-5p expression was reduced in NSCLC and was related to clinical parameters other than age and smoking, especially in the LUAD group.
Expression of miR-204-5p in non-small cell lung cancer (NSCLC) and subgroups derived from RT-qPCR and TCGA database. a Scatter plot for RT-qPCR indicated significantly lower miR-204-5p expression in NSCLC tissues (3.6760 ± 1.87670) than in the controls (4.6487 ± 2.46888) (P = 0.001). b Expression differences for miR-204-5p between lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) determined by RT-qPCR. The decrease was more obvious in LUAD (3.4663 ± 1.82397) than in LUSC (4.6870 ± 1.80638) (P = 0.006). c Expression level of miRNA-sequencing data from TCGA revealed significantly lower miR-204-5p expression in NSCLC tissues (1.8877 ± 2.18763) than in healthy control tissues (2.5944 ± 0.9404) (P = 0.000). d Expression comparison of miR-204-5p between LUAD (1.3331 ± 1.64315) and LUSC from TCGA (2.4922 ± 2.52336). The difference was consistent with the foregoing data of RT-qPCR. (P = 0.000)
Verification of miR-204-5p expression in TCGA
In this validation set, the levels of miR-204-5p were markedly decreased in NSCLC when compared to the normal control tissues (P = 0.000) (Fig. 2c). The TCGA Records were divided into TCGA-LUAD (containing 521 tumor cases and 46 controls) and TCGA-LUSC (478 tumor cases and 45 controls) due to the possibility of expression differences. The detected levels of miR-204-5p was lower in TCGA-LUAD, which was consistent with our results (P = 0.006) (Fig. 2d).
Results of data mining
Another 33 findings were selected for further analyses: 28 GEO datasets, 1 TCGA and 4 qualified publications. The first 11 investigations were involved monitoring of plasma samples, whereas the 22 analyzed solid tissues. The study by Guo W [15] was the only one derived from PubMed in this portion; papers 1 through 3 [28,29,30] were Chinese articles. The included datasets contained 3168 NSCLC cases and 1542 control samples. After acquisition of miRNA-204-5p from GEO, the means and SDs were calculated to assess its status in NSCLC. Detailed outcomes are listed in Table 4 and scatter point plots are presented in Fig. 3, 4.
Four GEO datasets assayed in plasma samples showed significant differences, but only two (GSE17681 and PMID:26497897) demonstrated the decreased level of miRNA-204-5p expression in NSCLC. Another 8 investigations performed in tissues were reflected in statistical significance except for the TCGA results; seven of these indicated a downregulation of miRNA-204-5p expression in NSCLC tissue specimens.
Integrated meta-analyses of miR-204-5p datasets in NSCLC
Each meta-analysis was first individually processed to evaluate level of miR-204-5p in the GEO data which covered 1747 NSCLC patients and 1105 control samples. Dysregulation of miR-204-5p was evident in NSCLC (SMD = − 0.098, 95% CI: − 0.310 to 0.114), but with poor statistical significance (P = 0.366) and high heterogeneity (I2 = 81.8%, P = 0.000) (Fig. 5a). Unexpected outcomes were obtained from subgroup meta-analysis, which indicated that the decrease was significantly different in both plasma (SMD = 0.374, 95% CI: 0.005 to 0.743, P = 0.047) and (SMD = − 0.098, 95% CI: − 0.310 to 0.114, P = 0.007) tissues, but also suggested a more sensitive response in cancerous tissues and evident heterogeneity (I2 > 90%, P = 0.000) as well (Fig. 5b). Random models were used to reduce the impact of heterogeneity.
An integrative meta-analysis of the entire data collection obtained from GEO, TCGA, publications and our RT-qPCR analyses was conducted to obtain a more precise assessment of miR-204-5p expression. Down-regulation of miR-204-5p in NSCLC (overall pooled SMD = − 0.447, 95% CI: − 0.750 ~ − 0.144, P = 0.004) was confirmed by the forest graph displayed in Fig. 6a, and the reduction was more significant in tissues (SMD = − 0.760, 95% CI: − 1.132 to − 0.378, P = 0.000) than in plasma (SMD = 0.224, 95% CI: − 0.301 to 0.749, P = 0.403) (Fig. 6b). Since substantial heterogeneity (I2 > 90%, P = 0.000) between data sources was noted between the data sources, a random model was adopted. The decline in miR-204-5p expression was more distinct in LUAD (SMD = − 0.258, 95% CI: − 0.685 to 0.169) than LUSC (SMD = − 0.012, 95% CI: − 0.406 to 0.382), though this subgroup analysis displayed weak statistical significance (P = 0.313) and considerable heterogeneity (I2 = 87.8%) (Fig. 6c). Furthermore, the reduction in miR-204-5p expression seemed more evident in LUAD tissues (SMD = − 0.554, 95% CI:-0.909 to − 0.199) than in plasma (SMD =1.176, 95% CI: − 0.397 to 2.748), but again the differences were not statistically significant (P = 0.236) and the data showed marked heterogeneity (I2 = 86.2%)(Fig. 6d).
Forest plot and subgroup analysis of miR-204-5p level in non-small cell lung cancer (NSCLC). a NSCLC vs. healthy controls; random-effects model. b Subgroup analysis layered by specimen source, plasma and tissue; random-effects model. c Subgroup analysis between lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC); random-effects model. d Subgroup analysis of LUAD sources, plasma and tissue; random-effects model
Clinical role of miR-204-5p in NSCLC
In total,31 records, which included 4368 samples derived from 28 GEO datasets, 1 TCGA, 1 publication and our study (Table 5), were used for diagnosis meta-analysis to survey the clinical role of miR-204-5p in NSCLC. Prior to the diagnosis meta-analysis, ROC curve for every case was generated and 4-fold table data were calculated. As showed in Figs. 7 and 8, the ROC curves presented varied diagnostic value with most of them revealing relatively high region in solid tissues, in agreement with TCGA and our study (Fig. 9).
The miR-204-5p diagnostic accuracy and its significance in NSCLC was further examined in SROC plots integrating all GEO datasets, TCGA, publications and our study to arrive at a reliable conclusion. Simultaneous subgroup analysis was conducted on the experimental sources and tumor types. The whole combined area under the curve (AUC) was 0.74 (95% CI: 0.70–0.77) with a sensitivity and specificity of 0.76 and 0.58 respectively (Fig. 10). The AUCs from different sample origins were similar to the combined AUC, whereas polarization of the sensitivity and specificity was evident in the plasma portion (Fig. 11). The AUC was larger for the entire LUAD group (0.78, 95% CI: 0.74–0.81) than for the entire LUSC group (0.66, 95% CI: 0.62–0.70), and showed higher sensitivity (0.63 to 0.32) and lower specificity (0.78 to 0.90). However, significant heterogeneity was evident by the large Q and I2 values, except in the LUSC subgroup (Table 6).
Prognostic evaluation of miR-204-5p in NSCLC
The K-M plots of our RT-qPCR data indicated a correlation between the NSCLC survival rate and miR-204-5p expression, as patients with higher levels of miR-204-5p survived longer than those with lower expression, although the difference did not meet statistical significance (Log Rank P = 0.231) (Fig. 12a).
Only two publications were deemed eligible for prognostic assessment. The general information of 2 included references, 3 GEO datasets, and our study matched the required assessment conditions for a sum of 415 participants, as shown in Table 7. The HR and 95% CI were not included in the paper by Shi L [59], so they were calculated from the K-M survival curves, and the results with high statistical significance was considered in the selection for next step.
The results shown in Fig. 12b and c indicate that the use of miR-204-5p as an auxiliary prognostic risk factor for NSCLC patients is not possible at present, due to the lack of statistical significance in the prognostic meta-analysis (95% CI: 0.660 to 1.188), and a lack of large-scale investigations in plasma.
Screening and validation of miR-204-5p target genes
In total, 4399 target genes were identified in at least six online predicted applications from miRwalk and 4371 up-regulated genes with FDR<0.05 were screened from TCGA. Subsequent analysis therefore focused on 541 over-active candidate genes from the intersection of miRwalk and TCGA.
The DAVID online tool identified 106 terms from the GO analysis and the top 3 most significantly enriched items associated with biological process (BP), cellular component (CC), and molecular function (MF) are listed in Table 8 (P < 0.05). The relevant target genes were chiefly involved in neuron projection, transcription factor activity, RNA polymerase II transcription regulation, extracellular matrix metabolism, and ion channel activity. In addition, 7 enriched pathways of KEGG analysis were collected from the same platform. As shown in Table 8, the top 3 signal pathways (P < 0.05) were connected with microRNAs in cancer, cell adhesion molecules (CAMs), and signaling pathways regulating pluripotency of stem cells.
Taking the differences in genetic expression and function into account, the PPI network of 117 DEGs from the top three GO items and KEGG pathways was explored by STRING and visualized by Cytoscape to determine the interaction between the proteins encoded by candidate target genes. As Fig. 13 shows, the network consisted of 117 nodes and 130 edges. The top 6 proteins with the highest degrees of connectivity were HDAC1 (degree = 10), SCN8A (degree = 9), DLG1 (degree = 8), EPHB2 (degree = 8), GDNF (degree = 8) and CALB1 (degree = 8).
Scatter point plots from GEPIA indicated that expression of the six hub genes was elevated in NSCLC, and that EPHB2 had the most apparent variation. Of particular interest, DLG1 and GDNF showed a pronounced trend of over-expression in LUSC (Fig. 14). Besides no record about SCN8A, THPA confirmed a similar tendency for an increased expression of HDAC1, DLG1, EPHB2 and CALB1(Fig. 15), while GDNF was not apparently changed in either normal or lung cancer tissues; no data were available for SCN8A. In addition to GDNF (aliases ATF or ATF2) identified from the available literature [60], EPHB2 and DLG1 have been proposed as suitable targets of miR-204-5p. Further comprehensive investigations and systematic evaluations are needed to confirm this hypothesis because of small sample size of THPA and a lack of statistical analysis.
Protein expression variation of hub genes in non-small cell lung cancer (NSCLC) tissues and normal pneumocytes from THPA. a HDAC1 in normal pneumocytes (antibody CAB068191). b HDAC1 in NSCLC tissues (antibody CAB068191). c DLG1 in normal pneumocytes (antibody CAB016307). d DLG1 in NSCLC tissues (antibody CAB016307). e EPHB2 in normal pneumocytes (antibody CAB013647). f EPHB2 in NSCLC tissues (antibody CAB013647). g GDNF in normal pneumocytes (antibody CAB005210). h GDNF in NSCLC tissues (antibody CAB005210). i: CALB1 in normal pneumocytes (antibody HPA023099). j CALB1 in NSCLC tissues (antibody HPA023099). From the immunohistochemistry results of THPA, HDAC1, DLG1, EPHB2 and CALB1 trended to raise in NSCLC tissues, but GDNF was not apparently changed. Beyond that, no data was available for SCN8A
TFs and the miR-204-5p regulatory network
In the present work, 61, 89, and 66 TFs related to miR-204-5p were obtained from GTRD, HTFtarget and Transmir, respectively. TF prediction was mainly matched examined for GDNF, DLG1, and EPHB2 since these genes were implicated as likely target genes. In total, GTRD and HTFtarget revealed 378 and 122 TFs of EPHB2, 408 and 171 TFs of DLG1, 271and 89 TFs with GDNF, respectively. The intersection outcome revealed MAX, MYC, and RUNX1 as the main TFs associated with miR-204-5p, GDNF, EPHB2, and DLG1(Fig. 16), while MAX was associated with miR-204-5p by TFBS prediction in JASPAR, which included some similar sequences in miR-204-5p, EPHB2, and GDNF. In addition, the putative TFBSs of MYC approached a certain degree of coincidence with MAX (Table 9). Binding competition of miRNA towards hub genes was confirmed by the miRwalk database and publication, but pairwise interactions of miR-204-5p with TFs and TFs interactions with hub genes could not be definitively constructed due to lack of database prediction, experimental proof and literature confirmation, so structural motifs of miR-204-5p networks could not be established (Fig. 17).
Venn charts for transcription factor screening. a Predicted transcription factors (TFs) of miR-204-5p from GTRD, HTF target and Transmir. b Predicted TFs of the three crucial hub genes from GTRD. c Predicted TFs of the three crucial hub genes from HTF target. d Predicted candidate TFs from coherent intersection of the foregoing three
Discussion
The data presented here verified the decrease in miR-204-5p expression in NSCLC by comprehensive analysis of RT-qPCR, microarrays, sequencing data, and publications and revealed an obvious decrease in cancerous tissues and the LUAD subtype. An auxiliary role for miR-204-5p was also identified by in NSCLC, particularly in tissues and LUAD, which was verified by the meta-analysis. Unfortunately, the prognostic implications for miR-204-5p were weak and showed no statistical significance in the meta-analysis, due to a shortage of large-scale investigations. Nevertheless, the information provided by GO annotation and KEGG analysis indicated that the target genes of miR-204-5p were associated with neuron projection, transcription factor activity, RNA polymerase II transcription regulation, extracellular matrix (ECM) metabolism, and ion channel activity, as well as were connected with microRNAs in cancer, cell adhesion molecules (CAMs) and signaling pathways regulating pluripotency of stem cells. Three of the six hub genes, GDNF, EPHB2 and DLG1, were selected for continued research due to their distinct characteristics in NSCLC. TF prediction revealed speculatively functional relations of MAX, MYC, and RUNX1 between miR-204-5p and these three genes, although only MAX demonstrated the TFBS sequences connected with miR-204-5p upon further query.
At present, miR-204-5p has aroused considerable interest in cancer research for its dual function as an oncogene and tumor suppressor [14]. MiR-204-5p is clearly attenuated in NSCLC, and its expression is negatively linked with tumor size, clinical stage, and metastasis [28,29,30, 59, 61]. Downregulation of miR-204 occurs in part due to its hypermethylation in the promoter region [59]. Elevated expression of miR-204-5p depresses NSCLC migration and invasion by targeting Janus kinase 2 (JAK2) [17], restrains proliferation of NSCLC cells by regulating SIX homeobox 1 (SIX1) and attenuates LUAD angiogenesis potentially by JAK2-signal transducer and activator of transcription 3 (JAK2-STAT3) pathway [16]. In addition, miR-204-5p serves as a cancer suppressor gene by modulating oncogenic Wnt/FZD signaling pathways [62], inhibiting NUAK family kinase 1 (NUAK1) in NSCLC [59], and mediating a long-noncoding RNA (lncRNA) MALAT1 effect on the epithelial-to-mesenchymal transition (EMT) and cells invasion [63]. Our results confirmed the downregulation of miR-204-5p expression in NSCLC and revealed a constant level of decline in LUAD.
The integrative meta-analysis also indicated a promising role for miR-204-5p for NSCLC screening, as did subgroup SROC curves, even though the sample origins were different. The variation in miR-204-5p expression in tissues was helpful in diagnosis of LUAD than of LUSC. Considering the invasive work, high cost, and cumbersome procedure of tissue biopsy, analysis of blood circulating miR-204-5p was considered an attractive screening indicator. However, low sensitivity, high specificity, and small sample sizes of the currently available data mean that more research and detailed profiling at all levels are needed to provide information to confirm the effectiveness of blood screening.
The correlation between the low miR-204-5p and high risk of death in patients with NSCLC [15, 59] was evident in our study but failed to reach statistical significance. The subsequent integrative meta-analysis was conducted to gain insights into the potential usefulness of miR-204-5p in NSCLC prognosis, but the data from the literature and from the present study regarding the ability of miR-204-5p to predict survival times of patients with NSCLC are conflicting. Consequently, no conclusion can be made in terms of miR-204-5p for NSCLC, at least for now.
The GO analysis indicated that the estimated target genes were mainly enriched in neuron projection, transcription factor activity, RNA polymerase II transcription regulation, ECM metabolism and ion channel activity, suggesting a potential involvement of miR-204-5p in the molecular function and signal modulation associated with NSCLC biological processes. The KEGG pathway analysis indicated that some of candidate genes were participated in microRNAs in cancer, CAMs, and signaling pathways regulating pluripotency of stem cells. Like other miRNAs, miR-204-5p plays an indispensable role in cancer proliferation, migration, and metastasis by regulating the tumor microenvironment, such as ECM structure and CAM metabolism [64]. Cancer stem cells (CSCs) attain stemness by complicated processes and signaling pathways, such as JAK-STAT, nuclear factor kappa B, Sonic hedgehog, transforming growth factor beta, Wnt/β-catenin, and PI3K/AKT [65, 66]. Many miRNAs take part in processes that maintain a balance between differentiation and quiescence of pulmonary CSCs, adjust the tumor microenvironment and affect cell cycle progression via regulation of these signaling pathways [67]. Consequently, miR-204-5p and its target genes could serve as important determinants of NSCLC pathogenesis and development.
Continued investigation of the hub genes involved in GO enrichment and KEGG Pathway analysis identified six leading relevant genes that were screened out due to binding to 3’UTR of miRNA. However, only GDNF has been investigated for a direct relationship with miR-204-5p in NSCLC [60].
GDNF, also called ATF or ATF2, is a well-characterized oncogene that promotes tumor growth, invasion, and metastasis, in addition to tumor microenvironment alterations [68]. GDNF expression occurs high level in NSCLC, though a significant difference exists with regard to factors such as race, gender, age, smoking status, and histologic subtype [69]. GDNF is upregulated at the transcript level in LUSC [70], and is hypermethylated in tumor tissues [71]. It also facilitates demethylation of the fibromodulin promoter and promotes subsequent angiogenesis in human glioblastomas [72]. Nerve-derived GDNF increases programmed death ligand 1 (PD-L1) levels in head and neck squamous cell carcinoma cells by activating the JAK2-STAT1 signaling pathway, which in turn promotes the evasion of cancer cells from immune system surveillance in the nerve-cancer microenvironment [73]. Recent research in colorectal cancer (CRC) has indicated that miR-196a-5p exerts its function in cell proliferation and migration by regulating GDNF expression [74], while miR-451 influences drug resistance in renal cell carcinoma by targeting GDNF [75]. GDNF is also targeted and regulated by miR-204-5p which inversely affects GDNF mRNA and protein levels, to inhibit NSCLC growth, migration, and cell cycle alteration and promote apoptosis [60]. Therefore, the interaction between miR-204-5p and GDNF appears to be critical in the development and progression of NSCLC and requires thorough research.
DLG1 is a vital participant in the control of cellular processes like polarity, proliferation and migration, so its dysregulation and mutation give rise to pathologies that include oncogenic processes [76]. DLG1 is mainly identified as a tumor suppressor, since overexpression is observed early in the onset of cervical cancer (CeCa) [77] and elevated DLG1 promotes intestinal tumorigenesis [78], predicts poor prognosis in people with CRC [79] and increases the invasiveness of NSCLC cell lines [80]. Increased phosphorylation of the DLG1 SH3-Hook region promotes interaction with the PDZ ligand of PKCα and accelerates cell migration [80]. The lncRNA DLG1-AS1 acts as a competitive inhibitor that influences the activity of miR-107 on its target gene ZHX1, thereby inducing cancer cell proliferation [81]. Moreover, DLG1 deficiency results in incorrect spindle polarity and a delay in cells transiting orientation [78], which disrupts cellular structure and distribution [82]. Interestingly, DLG1 protein levels are significantly lower in NSCLC and hepatocellular carcinoma (HCC) than in the corresponding normal tissues [83, 84], but are nearly undetectable in poorly differentiated stages of colon adenocarcinoma [85], in contrast to our findings and the existing literature. One possible reason is that DLG1 dysregulation in advanced tumor progression or in more malignant forms depends on its spatial/temporal distribution. Future research should focus on this possibility.
A series of studies have reported a direct correlation between EPHB2 expression and numerous human malignancies, including NSCLC. EPHB2 activates bidirectional signaling cascades and its upregulation predicts poor survival in LUAD [86], CRC [87], breast cancer [88] and malignant mesothelioma [89]. One study indicated that EPHB2 enhances cellular growth, migration and invasion in CeCa by a competitive inhibition that counteracts the miR-204 effect on cell cycle arrest, Bax overexpression and PI3K/AKT signaling pathway deactivation via competitive inhibition [90]. Expression of miRNAs also significantly suppresses EPHB2 expression, resulting in a decrease of tubulogenesis and angiogenesis [91, 92]. EPHB2 affects cell viability in medulloblastoma in part by promotion of the G2/M phase of the cell cycle [93]. Activation of EPHB2 promotes the progression of cutaneous squamous cell carcinoma cells by accelerating the production of invasive proteinases like MMP13 and MMP1 [94]. Nevertheless, some publications highlight EPHB2 declines in CRC, which was supposedly attributable to EMT modulation [95] and epigenetic modification of promoter [96, 97]. Future research could identify where whether specific differences in the interaction of EPHB2 and miR-204-5p are associated with NSCLC.
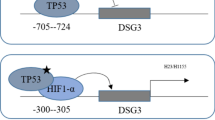
As discussed above, the function of miR-204-5p in NSCLC is also influenced by TFs as well, but an unanswered question is whether TFs regulate miR-204-5p or whether TFs could be adjusted or controlled by this miRNA in some way. Although great achievements have been made in understanding the biological behavior of miR-204-5p and its mRNA targets, integrative analysis of miRNA-TF-gene regulatory networks is still needed, as TFs are undoubtedly involved in pulmonary cancer initiation, progression, dissemination, recurrence, and even drug resistance [98]. At least ten kinds of TF-miRNA synergistic regulatory networks apparently function in NSCLC [99]. In addition to combining with and regulating its target genes, miR-204-5p also attenuates some angiogenic inducers like hypoxia inducible factor-1α (HIF-1α) to impair angiogenesis in LUAD [16]. Another study has demonstrated a dependence of miR-204-5p level on promoter hypermethylation and support by positive feedback of three TFs, c-MYB, ETS1 and RUNX2 [100]. Osterix, a transcription factor that is essential and specific for osteogenesis, coordinately modulates miR-204-5p and its endogenous competitors, as well as ultimately establishing a feed-forward loop (FFL) ultimately [101]. Activation of STAT3 suppresses miR-204-5p activities, in turn affecting proliferation and apoptotic resistance in human pulmonary arterial hypertension and nasopharyngeal carcinoma [102, 103]. A positive FFL between Hepatitis B virus, miR-204-5p, and STAT3 appears to contribute to HCC incidence [104].
Other work has suggested that miR-204-5p reciprocally represses TrkB expression; however, TrkB expression noticeably increases JAK2 and STAT3 phosphorylation. The phospho-STAT3 then directly binds to promoter sequence of miR-204-5p, resulting in increased clonogenic proliferation, migration and invasion in endometrial carcinoma, via a feed-backward-loop (FBL) motif of TF-miRNA-target gene [105]. The activity of the IL-6R/STAT3/miR-204-5p FBL also leads to chemosensitivity [106]. The possibility exists that, at molecular level, the cellular activities involved in NSCLC progression, including tumor cell proliferation, differentiation, invasion, apoptosis, recurrence, and even drug resistance, are associated with downregulation of miR-204-5p and account for the direct upregulation of its target genes as well as unidirectional or bidirectional activation of TFs.
Among the three TFs expected to take part in miR-204-5p networks, MAX forms a dimer-complex system of transcriptional regulation with other family members, which include MYC, Mad and Mxi1, and is implicated in cell proliferation, differentiation and apoptosis [107]. The sequence similarity in MAX and MYC TFBS in the current work was predicted based on the existence of the multiprotein complex. An increased expression of miR-22 in leukemia cells reduces the MAX expression level, blocking cell cycle progression at the G1 phase [108]. MAX expression in HCC activates Linc00176, which is a competing endogenous lncRNA (ceRNA) of tumor-suppressive miRNA, resulting in cell cycle acceleration and reduction of apoptosis by reducing the levels of miR-9 and miR-185 [109]. In CRC, a MAX/MYC heterodimer induced by elevated HIF-2α mediates transcriptional repression of hypoxia-related miR-15-16, leading to tumor angiogenesis and hematogenous metastasis by further loss of post-transcriptional restriction towards fibroblast growth factor-2 [110]. MYC, also known as MYCC and c-Myc, is frequently amplified in numerous human cancers via transcriptional regulation of specific target genes, including miRNA and lncRNA [111]. MiR-296 − 3p directly targets PRKCA to impair FAK-Ras-MYC signaling, thereby accelerating its own transcription in a FBL that obstructs the EMT signal and progression through the cell cycle, following suppression of cell proliferation, metastasis and chemosensitivity in LUAD [112]. Another study has suggested that miR-342-3p is capable of indirectly adjusting MYC by directly repressing E2F1, a MYC-collaborating molecule [52]. In breast tumor, MYC expression correlates positively with miR-203b-3p and miR-203a-3p but negatively with BCL2L1 expression, resulting in formation of a TF-FFL [113]. MIR7-3HG restrains MYC dephosphorylation by downregulation of AMBRA1 to form a positive feedback loop for its own expression and further contributing significantly to autophagic control [114].
As a crucial hematopoietic transcription factor, RUNX1 is well-documented in chromosomal translocations and in several types of carcinogenesis processes [115]. RUNX1 is positioned in the center of miRNA circuits relevant for malignant hematopoiesis in transcriptional programs [116, 117]. A RUNX1-microRNA-139-HCP5 axis shows a positive FBL for mediating the tumor-suppressive effects of glioma cells [118]. A miR-18a-RUNX1-ZO-1 regulatory network also increases the permeability of the blood-tumor barrier (BTB), thereby providing novel potential targets for drug transportation across the BTB as an attractive strategy for glioma treatment [119]. By binding to the miRNA promoter, RUNX1 increases the transcriptional level of miR-27a in breast cancer and concomitantly the decreases expression of ZBTB10, a direct target gene of miR-27a, to promote endothelial differentiation and subsequent angiogenesis and tumor metastasis [120]. Conversely, reduced expression of Runx1 in breast cancer cells leads to elevated expression of both pre-miR-378 and PPARGC1B, which is a host gene of miR-378, to create a FBL on that reduces cell migration and invasion [121]. The miR-204-5p circuits and its hub genes and TFs still await identification, but TFBS prediction was capable of offering fresh perspectives, and likewise, assisting in new theoretical insights into potential regulatory mechanisms. Based on the available data, MAX would appear likely to be a vital TF involved in miR-204-5p-mRNA interactions, since it was the only assumed attachment that focused on the upstream region of genetic sequences in the current work.
This study had drawbacks and limitations. One limitation is that the findings indicate a great heterogeneity between the data sources, which were then explored via a random effects model and subgroup meta-analyses. The trial quality was also generally poor due to heterogeneity that remained above 50%. One possible cause of the statistical heterogeneity is that the data were generated using different sources, operating protocols, and detection metrics. Univariate survival analysis is also the only appraisal method for determining the prognostic significance of miR-204-5p. A carefully designed evaluation system should be developed to provide a more in-depth assessment of this issue. In particular, time limits and tight budgets have prevented a satisfactory generation of experimental proof to validate the function of miR-204-5p regulatory networks with the target genes and TFs. In addition, the biological progression and molecular regulation of NSCLC is complicated, so other mechanisms mediated by miRNA circuits should also be addressed with in-depth research.
Conclusion
As the most frequent type of pulmonary cancer, NSCLC deserves more effort in achieving the goal of early detection and timely treatment. The findings presented in the current research demonstrated an attenuation of miR-204-5p expression in NSCLC, this decrease was more frequently observed in cancerous tissues and in the LUAD subtype and was, in part, helpful for diagnosis. The activities of miR-204-5p as an anti-oncogene were induced by its regulatory axes or circuits with target genes and TFs that participated in specific genetic pathways and biological processes.
Availability of data and materials
Data and material will be available on reasonable request.
Abbreviations
- 95% CI:
-
95% confidence interval
- DEGs:
-
Differentially expressed genes
- GEO:
-
Gene expression omnibus
- GO:
-
Gene ontology
- HR:
-
Hazard ratio
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- K-M curve:
-
Kaplan-Meier curve
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- MiRNA:
-
microRNA
- NSCLC:
-
Non-small cell lung cancer
- ROC:
-
Receiver operating characteristic
- RT-qPCR:
-
Real-time quantitative PCR
- SMD:
-
Standard mean deviation
- TCGA:
-
The cancer genome atlas
- TF:
-
Transcription factor
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34.
Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–53.
Smolle E, Pichler M. Non-smoking-associated lung cancer: a distinct entity in terms of tumor biology, patient characteristics and impact of hereditary Cancer predisposition. Cancers (Basel). 2019; -https://doi.org/10.3390/cancers11020204.
Luo W, Rao M, Qu J, Luo D. Applications of liquid biopsy in lung cancer-diagnosis, prognosis prediction, and disease monitoring. Am J Transl Res. 2018;10:3911–23.
Adderley H, Blackhall FH, Lindsay CR. KRAS-mutant non-small cell lung cancer: converging small molecules and immune checkpoint inhibition. EBioMedicine. 2019; -https://doi.org/10.1016/j.ebiom.2019.02.049.
Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. 2019; -https://doi.org/10.5334/aogh.2419.
Conway FM, Garner JL, Orton CM, Srikanthan K, Kemp SV, Shah PL. Contemporary concise review 2018: lung cancer and pleural disease. Respirology. 2019;24:475–83.
Qin H, Wang F, Liu H, Zeng Z, Wang S, Pan X, Gao H. New advances in immunotherapy for non-small cell lung cancer. Am J Transl Res. 2018;10:2234–45.
Rong B, Yang S. Molecular mechanism and targeted therapy of Hsp90 involved in lung cancer: new discoveries and developments (review). Int J Oncol. 2018;52:321–36.
Ors-Kumoglu G, Gulce-Iz S, Biray-Avci C. Therapeutic microRNAs in human cancer. Cytotechnology. 2019;71:411–25.
Hu X, Liao S, Bai H, Wu L, Wang M, Wu Q, Zhou J, Jiao L, Chen X, Zhou Y, et al. Integrating exosomal microRNAs and electronic health data improved tuberculosis diagnosis. EBioMedicine. 2019;40:564–73.
McCubrey JA, Lertpiriyapong K, Steelman LS, Abrams SL, Yang LV, Murata RM, Rosalen PL, Scalisi A, Neri LM, Cocco L, et al. Effects of resveratrol, curcumin, berberine and other nutraceuticals on aging, cancer development, cancer stem cells and microRNAs. Aging (Albany NY). 2017;9:1477–536.
Chen L, Sun H, Wang C, Yang Y, Zhang M, Wong G. miRNA arm switching identifies novel tumour biomarkers. EBioMedicine. 2018;38:37–46.
Li T, Pan H, Li R. The dual regulatory role of miR-204 in cancer. Tumour Biol. 2016;37:11667–77.
Guo W, Zhang Y, Zhang Y, Shi Y, Xi J, Fan H, Xu S. Decreased expression of miR-204 in plasma is associated with a poor prognosis in patients with non-small cell lung cancer. Int J Mol Med. 2015;36:1720–6.
Liu X, Gao X, Zhang W, Zhu T, Bi W, Zhang Y. MicroRNA-204 deregulation in lung adenocarcinoma controls the biological behaviors of endothelial cells potentially by modulating Janus kinase 2-signal transducer and activator of transcription 3 pathway. IUBMB Life. 2018;70:81–91.
Wang P, Lv HY, Zhou DM, Zhang EN. miR-204 suppresses non-small-cell lung carcinoma (NSCLC) invasion and migration by targeting JAK2. Genet Mol Res. 2016;15.
Butnor KJ, Beasley MB, Cagle PT, Grunberg SM, Kong FM, Marchevsky A, Okby NT, Roggli VL, Suster S, Tazelaar HD, Travis WD. Protocol for the examination of specimens from patients with primary non-small cell carcinoma, small cell carcinoma, or carcinoid tumor of the lung. Arch Pathol Lab Med. 2009;133:1552–9.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–8.
He R, Gao L, Ma J, Peng Z, Zhou S, Yang L, Feng Z, Dang Y, Chen G. The essential role of MTDH in the progression of HCC: a study with immunohistochemistry, TCGA, meta-analysis and in vitro investigation. Am J Transl Res. 2017;9:1561–79.
Ye ZH, Wen DY, Cai XY, Liang L, Wu PR, Qin H, Yang H, He Y, Chen G. The protective value of miR-204-5p for prognosis and its potential gene network in various malignancies: a comprehensive exploration based on RNA-seq high-throughput data and bioinformatics. Oncotarget. 2017;8:104960–80.
Gao L, Li SH, Tian YX, Zhu QQ, Chen G, Pang YY, Hu XH. Role of downregulated miR-133a-3p expression in bladder cancer: a bioinformatics study. Onco Targets Ther. 2017;10:3667–83.
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60.
Melsen WG, Bootsma MC, Rovers MM, Bonten MJ. The effects of clinical and statistical heterogeneity on the predictive values of results from meta-analyses. Clin Microbiol Infect. 2014;20:123–9.
Schou IM, Marschner IC. Meta-analysis of clinical trials with early stopping: an investigation of potential bias. Stat Med. 2013;32:4859–74.
Li LX, LD, YYL, Chen M, Lu QC, Wu D. Expression of miR-204 in non-small cell lung cancer and its targeted regulation on SIRT1. J Guangdong Med Univ. 2017;35:266–70.
Wang QC, WQ, Wan LX, Qu ZY, Zhao DB, Zhu K. microRNA-204 expression in non-small cell lung cancer and its effcts on H252 cells proliferation and apoptosis. Chin J Lab Diagn. 2018;22:1009–12.
Xu YZ, TZ, LC. miRNA-204 expression in non-small cell lung cancer tissue and its effect on cancer cell proliferation and apoptosis. China J Modern Med. 2018;28:57–61.
Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4:e6229.
Keller A, Leidinger P, Borries A, Wendschlag A, Wucherpfennig F, Scheffler M, Huwer H, Lenhof HP, Meese E. miRNAs in lung cancer - studying complex fingerprints in patient's blood cells by microarray experiments. BMC Cancer. 2009;9:353.
Keller ALP. Peripheral profiles from patients with cancerous and non cancerous lung diseases. Gene Expr Omnibus database. 2010; -https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24709. Accessed 31 Oct 2018.
Patnaik SK, Yendamuri S, Kannisto E, Kucharczuk JC, Singhal S, Vachani A. MicroRNA expression profiles of whole blood in lung adenocarcinoma. PLoS One. 2012;7:e46045.
Keller A, Leidinger P, Bauer A, Elsharawy A, Haas J, Backes C, Wendschlag A, Giese N, Tjaden C, Ott K, et al. Toward the blood-borne miRNome of human diseases. Nat Methods. 2011;8:841–3.
Patnaik SK, Kannisto ED, Mallick R, Vachani A, Yendamuri S. Whole blood microRNA expression may not be useful for screening non-small cell lung cancer. PLoS One. 2017;12:e0181926.
Godrey A, XZ, Sieber S, Hayes N, Rivera P, Taylor J. Serum miRNA expression is associated with lung cancer in a case-control study and normalizes following surgical resection. Gene Expr Omnibus Database. 2013; -https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46729. Accessed 31 Oct 2018.
Keller A, Leidinger P, Vogel B, Backes C, ElSharawy A, Galata V, Mueller SC, Marquart S, Schrauder MG, Strick R, et al. miRNAs can be generally associated with human pathologies as exemplified for miR-144. BMC Med. 2014;12:224.
Leidinger P, Galata V, Backes C, Stahler C, Rheinheimer S, Huwer H, Meese E, Keller A. Longitudinal study on circulating miRNAs in patients after lung cancer resection. Oncotarget. 2015;6:16674–85.
Liu X, Qu L. Gene expression-signatures for non-small cell lung cancer patients with different EGFR muational status. Gene Expr Omnibus Database. 2017; -https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93300. Accessed 31 Oct 2018.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8.
Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–90.
Tan X, Qin W, Zhang L, Hang J, Li B, Zhang C, Wan J, Zhou F, Shao K, Sun Y, et al. A 5-microRNA signature for lung squamous cell carcinoma diagnosis and hsa-miR-31 for prognosis. Clin Cancer Res. 2011;17:6802–11.
Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer DG. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–83.
Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K, Cardinaud B, Hofman V, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–78.
Ohba T, Nagano H. A small-cell lung cancer subtype with good prognosis found by a three miRNA signature. Gene Expr Omnibus Database. 2010; -https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19945. Accessed 31 Oct 2018.
Nymark P, Guled M, Borze I, Faisal A, Lahti L, Salmenkivi K, Kettunen E, Anttila S, Knuutila S. Integrative analysis of microRNA, mRNA and aCGH data reveals asbestos- and histology-related changes in lung cancer. Genes Chromosom Cancer. 2011;50:585–97.
Ma L, Huang Y, Zhu W, Zhou S, Zhou J, Zeng F, Liu X, Zhang Y, Yu J. An integrated analysis of miRNA and mRNA expressions in non-small cell lung cancers. PLoS One. 2011;6:e26502.
Jang JS, Jeon HS, Sun Z, Aubry MC, Tang H, Park CH, Rakhshan F, Schultz DA, Kolbert CP, Lupu R, et al. Increased miR-708 expression in NSCLC and its association with poor survival in lung adenocarcinoma from never smokers. Clin Cancer Res. 2012;18:3658–67.
van Jaarsveld MT, Wouters MD, Boersma AW, Smid M, van Ijcken WF, Mathijssen RH, Hoeijmakers JH, Martens JW, van Laere S, Wiemer EA, Pothof J. DNA damage responsive microRNAs misexpressed in human cancer modulate therapy sensitivity. Mol Oncol. 2014;8:458–68.
Bjaanaes MM, Halvorsen AR, Solberg S, Jorgensen L, Dragani TA, Galvan A, Colombo F, Anderlini M, Pastorino U, Kure E, et al. Unique microRNA-profiles in EGFR-mutated lung adenocarcinomas. Int J Cancer. 2014;135:1812–21.
Tai MC, Kajino T, Nakatochi M, Arima C, Shimada Y, Suzuki M, Miyoshi H, Yatabe Y, Yanagisawa K, Takahashi T. miR-342-3p regulates MYC transcriptional activity via direct repression of E2F1 in human lung cancer. Carcinogenesis. 2015;36:1464–73.
Pu HY, Xu R, Zhang MY, Yuan LJ, Hu JY, Huang GL, Wang HY. Identification of microRNA-615-3p as a novel tumor suppressor in non-small cell lung cancer. Oncol Lett. 2017;13:2403–10.
Fujita Y, Yagishita S, Hagiwara K, Yoshioka Y, Kosaka N, Takeshita F, Fujiwara T, Tsuta K, Nokihara H, Tamura T, et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther. 2015;23:717–27.
Robles AI, Arai E, Mathe EA, Okayama H, Schetter AJ, Brown D, Petersen D, Bowman ED, Noro R, Welsh JA, et al. An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J Thorac Oncol. 2015;10:1037–48.
Gasparini P, Cascione L, Landi L, Carasi S, Lovat F, Tibaldi C, Ali G, D'Incecco A, Minuti G, Chella A, et al. microRNA classifiers are powerful diagnostic/prognostic tools in ALK-, EGFR-, and KRAS-driven lung cancers. Proc Natl Acad Sci U S A. 2015;112:14924–9.
Jin Y, LY, Lu SH. The miRNA expression profiles in three subtypes of lung carcinomas. Gene Expr Omnibus Database. 2015; -https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74190. Accessed 31 Oct 2018.
Mitchell KA, Zingone A, Toulabi L, Boeckelman J, Ryan BM. Comparative Transcriptome profiling reveals coding and noncoding RNA differences in NSCLC from African Americans and European Americans. Clin Cancer Res. 2017;23:7412–25.
Shi L, Zhang B, Sun X, Lu S, Liu Z, Liu Y, Li H, Wang L, Wang X, Zhao C. MiR-204 inhibits human NSCLC metastasis through suppression of NUAK1. Br J Cancer. 2014;111:2316–27.
Zhang S, Gao L, Thakur A, Shi P, Liu F, Feng J, Wang T, Liang Y, Liu JJ, Chen M, Ren H. miRNA-204 suppresses human non-small cell lung cancer by targeting ATF2. Tumour Biol. 2016;37:11177–86.
Xia Y, Zhu Y, Ma T, Pan C, Wang J, He Z, Li Z, Qi X, Chen Y. miR-204 functions as a tumor suppressor by regulating SIX1 in NSCLC. FEBS Lett. 2014;588:3703–12.
Lin J, Zandi R, Shao R, Gu J, Ye Y, Wang J, Zhao Y, Pertsemlidis A, Wistuba II, Wu X, et al. A miR-SNP biomarker linked to an increased lung cancer survival by miRNA-mediated down-regulation of FZD4 expression and Wnt signaling. Sci Rep. 2017;7:9029.
Li J, Wang J, Chen Y, Li S, Jin M, Wang H, Chen Z, Yu W. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am J Cancer Res. 2016;6:1099–107.
Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Asp Med. 2018; -https://doi.org/10.1016/j.mam.2018.07.003.
Toledo-Guzman ME, Bigoni-Ordonez GD, Ibanez Hernandez M, Ortiz-Sanchez E. Cancer stem cell impact on clinical oncology. World J Stem Cells. 2018;10:183–95.
Heng WS, Gosens R, Kruyt FAE. Lung cancer stem cells: origin, features, maintenance mechanisms and therapeutic targeting. Biochem Pharmacol. 2019;160:121–33.
Fan T, Wang W, Zhang B, Xu Y, Chen L, Pan S, Hu H, Geng Q. Regulatory mechanisms of microRNAs in lung cancer stem cells. Springerplus. 2016;5:1762.
Mulligan LM. GDNF and the RET receptor in Cancer: new insights and therapeutic potential. Front Physiol. 2018;9:1873.
Lin C, Wang S, Xie W, Chang J, Gan Y. The RET fusion gene and its correlation with demographic and clinicopathological features of non-small cell lung cancer: a meta-analysis. Cancer Biol Ther. 2015;16:1019–28.
Garnis C, Davies JJ, Buys TP, Tsao MS, MacAulay C, Lam S, Lam WL. Chromosome 5p aberrations are early events in lung cancer: implication of glial cell line-derived neurotrophic factor in disease progression. Oncogene. 2005;24:4806–12.
Anglim PP, Galler JS, Koss MN, Hagen JA, Turla S, Campan M, Weisenberger DJ, Laird PW, Siegmund KD, Laird-Offringa IA. Identification of a panel of sensitive and specific DNA methylation markers for squamous cell lung cancer. Mol Cancer. 2008;7:62.
Chen M, Ba H, Lu C, Dai J, Sun J. Glial cell line-derived Neurotrophic factor (GDNF) promotes angiogenesis through the Demethylation of the Fibromodulin (FMOD) promoter in Glioblastoma. Med Sci Monit. 2018;24:6137–43.
Lin C, Cao W, Ren Z, Tang Y, Zhang C, Yang R, Chen Y, Liu Z, Peng C, Wang L, et al. GDNF secreted by nerves enhances PD-L1 expression via JAK2-STAT1 signaling activation in HNSCC. Oncoimmunology. 2017;6:e1353860.
Zhong F, Zhang W, Cao Y, Wen Q, Cao Y, Lou B, Li J, Shi W, Liu Y, Luo R, Chen C. LncRNA NEAT1 promotes colorectal cancer cell proliferation and migration via regulating glial cell-derived neurotrophic factor by sponging miR-196a-5p. Acta Biochim Biophys Sin Shanghai. 2018;50:1190–9.
Sun X, Lou L, Zhong K, Wan L. MicroRNA-451 regulates chemoresistance in renal cell carcinoma by targeting ATF-2 gene. Exp Biol Med (Maywood). 2017;242:1299–305.
Gardiol D, Marziali F, Dizanzo MP, Cavatorta AL. Differential expression of DLG1 as a common trait in different human diseases: an encouraging issue in molecular pathology. Biol Chem. 2018; -https://doi.org/10.1515/hsz-2018-0350.
Cavatorta AL, Di Gregorio A, Bugnon Valdano M, Marziali F, Cabral M, Bottai H, Cittadini J, Nocito AL, Gardiol D. DLG1 polarity protein expression associates with the disease progress of low-grade cervical intraepithelial lesions. Exp Mol Pathol. 2017;102:65–9.
Young MA, May S, Damo A, Yoon YS, Hur MW, Swat W, Parry L. Epigenetic regulation of Dlg1, via Kaiso, alters mitotic spindle polarity and promotes intestinal tumorigenesis. Mol Cancer Res. 2018; -https://doi.org/10.1158/1541-7786.mcr-18-0280.
Zhu GD, OuYang S, Liu F, Zhu ZG, Jiang FN, Zhang B. Elevated expression of DLG1 is associated with poor prognosis in patients with colorectal Cancer. Ann Clin Lab Sci. 2017;47:657–62.
O'Neill AK, Gallegos LL, Justilien V, Garcia EL, Leitges M, Fields AP, Hall RA, Newton AC. Protein kinase Calpha promotes cell migration through a PDZ-dependent interaction with its novel substrate discs large homolog 1 (DLG1). J Biol Chem. 2011;286:43559–68.
Rui X, Xu Y, Huang Y, Ji L, Jiang X. lncRNA DLG1-AS1 promotes cell proliferation by competitively binding with miR-107 and up-regulating ZHX1 expression in cervical Cancer. Cell Physiol Biochem. 2018;49:1792–803.
Marziali F, Bugnon Valdano M, Brunet Avalos C, Moriena L, Cavatorta AL, Gardiol D. Interference of HTLV-1 tax protein with cell polarity regulators: defining the subcellular localization of the tax-DLG1 interaction. Viruses. 2017;9:e355.
Szymanowska-Narloch A, Jassem E, Skrzypski M, Muley T, Meister M, Dienemann H, Taron M, Rosell R, Rzepko R, Jarzab M, et al. Molecular profiles of non-small cell lung cancers in cigarette smoking and never-smoking patients. Adv Med Sci. 2013;58:196–206.
Wu D, Liu G, Liu Y, Saiyin H, Wang C, Wei Z, Zen W, Liu D, Chen Q, Zhao Z, et al. Zinc finger protein 191 inhibits hepatocellular carcinoma metastasis through discs large 1-mediated yes-associated protein inactivation. Hepatology. 2016;64:1148–62.
Gardiol D, Zacchi A, Petrera F, Stanta G, Banks L. Human discs large and scrib are localized at the same regions in colon mucosa and changes in their expression patterns are correlated with loss of tissue architecture during malignant progression. Int J Cancer. 2006;119:1285–90.
Zhao C, Wang A, Lu F, Chen H, Fu P, Zhao X, Chen H. Overexpression of junctional adhesion molecule-a and EphB2 predicts poor survival in lung adenocarcinoma patients. Tumour Biol. 2017;39:1010428317691000.
Jang BG, Kim HS, Chang WY, Bae JM, Kang GH. Prognostic significance of EPHB2 expression in colorectal Cancer progression. J Pathol Transl Med. 2018;52:298–306.
Husa AM, Magic Z, Larsson M, Fornander T, Perez-Tenorio G. EPH/ephrin profile and EPHB2 expression predicts patient survival in breast cancer. Oncotarget. 2016;7:21362–80.
Goparaju C, Donington JS, Hsu T, Harrington R, Hirsch N, Pass HI. Overexpression of EPH receptor B2 in malignant mesothelioma correlates with oncogenic behavior. J Thorac Oncol. 2013;8:1203–11.
Duan S, Wu A, Chen Z, Yang Y, Liu L, Shu Q. miR-204 regulates cell proliferation and invasion by targeting EphB2 in human cervical Cancer. Oncol Res. 2018;26:713–23.
Biyashev D, Veliceasa D, Topczewski J, Topczewska JM, Mizgirev I, Vinokour E, Reddi AL, Licht JD, Revskoy SY, Volpert OV. miR-27b controls venous specification and tip cell fate. Blood. 2012;119:2679–87.
Keung MH, Chan LS, Kwok HH, Wong RN, Yue PY. Role of microRNA-520h in 20(R)-ginsenoside-Rg3-mediated angiosuppression. J Ginseng Res. 2016;40:151–9.
Bhatia S, Hirsch K, Bukkapatnam S, Baig NA, Oweida A, Griego A, Calame D, Sharma J, Donson A, Foreman N, et al. Combined EphB2 receptor knockdown with radiation decreases cell viability and invasion in medulloblastoma. Cancer Cell Int. 2017;17:41.
Farshchian M, Nissinen L, Siljamaki E, Riihila P, Toriseva M, Kivisaari A, Ala-Aho R, Kallajoki M, Verajankorva E, Honkanen HK, et al. EphB2 promotes progression of cutaneous squamous cell carcinoma. J Invest Dermatol. 2015;135:1882–92.
Schnappauf O, Beyes S, Dertmann A, Freihen V, Frey P, Jagle S, Rose K, Michoel T, Grosschedl R, Hecht A. Enhancer decommissioning by Snail1-induced competitive displacement of TCF7L2 and down-regulation of transcriptional activators results in EPHB2 silencing. Biochim Biophys Acta. 2016;1859:1353–67.
Alazzouzi H, Davalos V, Kokko A, Domingo E, Woerner SM, Wilson AJ, Konrad L, Laiho P, Espin E, Armengol M, et al. Mechanisms of inactivation of the receptor tyrosine kinase EPHB2 in colorectal tumors. Cancer Res. 2005;65:10170–3.
Lian H, Jia X, Shi N, Xie S, Wang J, Wang W, Ma F, Liu H, Wang A, Cheng X, Liu C. Notch signaling promotes serrated neoplasia pathway in colorectal cancer through epigenetic modification of EPHB2 and EPHB4. Cancer Manag Res. 2018;10:6129–41.
Slawek S, Szmyt K, Fularz M, Dziudzia J, Boruczkowski M, Sikora J, Kaczmarek M. Pluripotency transcription factors in lung cancer-a review. Tumour Biol. 2016;37:4241–9.
Li K, Li Z, Zhao N, Xu Y, Liu Y, Zhou Y, Shang D, Qiu F, Zhang R, Chang Z, Xu Y. Functional analysis of microRNA and transcription factor synergistic regulatory network based on identifying regulatory motifs in non-small cell lung cancer. BMC Syst Biol. 2013;7:122.
Todorova K, Metodiev MV, Metodieva G, Zasheva D, Mincheff M, Hayrabedyan S. miR-204 is dysregulated in metastatic prostate cancer in vitro. Mol Carcinog. 2016;55:131–47.
Chen Q, Liu W, Sinha KM, Yasuda H, de Crombrugghe B. Identification and characterization of microRNAs controlled by the osteoblast-specific transcription factor Osterix. PLoS One. 2013;8:e58104.
Courboulin A, Paulin R, Giguere NJ, Saksouk N, Perreault T, Meloche J, Paquet ER, Biardel S, Provencher S, Cote J, et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–48.
Ma L, Deng X, Wu M, Zhang G, Huang J. Down-regulation of miRNA-204 by LMP-1 enhances CDC42 activity and facilitates invasion of EBV-associated nasopharyngeal carcinoma cells. FEBS Lett. 2014;588:1562–70.
Huang JY, Chen HL, Shih C. MicroRNA miR-204 and miR-1236 inhibit hepatitis B virus replication via two different mechanisms. Sci Rep. 2016;6:34740.
Bao W, Wang HH, Tian FJ, He XY, Qiu MT, Wang JY, Zhang HJ, Wang LH, Wan XP. A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol Cancer. 2013;12:155.
Zhu X, Shen H, Yin X, Long L, Chen X, Feng F, Liu Y, Zhao P, Xu Y, Li M, et al. IL-6R/STAT3/miR-204 feedback loop contributes to cisplatin resistance of epithelial ovarian cancer cells. Oncotarget. 2017;8:39154–66.
Carroll PA, Freie BW, Mathsyaraja H, Eisenman RN. The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis. Front Med. 2018;12:412–25.
Ting Y, Medina DJ, Strair RK, Schaar DG. Differentiation-associated miR-22 represses max expression and inhibits cell cycle progression. Biochem Biophys Res Commun. 2010;394:606–11.
Tran DDH, Kessler C, Niehus SE, Mahnkopf M, Koch A, Tamura T. Myc target gene, long intergenic noncoding RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and cell survival by titrating tumor suppressor microRNAs. Oncogene. 2018;37:75–85.
Xue G, Yan HL, Zhang Y, Hao LQ, Zhu XT, Mei Q, Sun SH. c-Myc-mediated repression of miR-15-16 in hypoxia is induced by increased HIF-2alpha and promotes tumor angiogenesis and metastasis by upregulating FGF2. Oncogene. 2015;34:1393–406.
Swier L, Dzikiewicz-Krawczyk A, Winkle M, van den Berg A, Kluiver J. Intricate crosstalk between MYC and non-coding RNAs regulates hallmarks of cancer. Mol Oncol. 2019;13:26–45.
Fu Q, Song X, Liu Z, Deng X, Luo R, Ge C, Li R, Li Z, Zhao M, Chen Y, et al. miRomics and Proteomics Reveal a miR-296-3p/PRKCA/FAK/Ras/c-Myc Feedback Loop Modulated by HDGF/DDX5/beta-catenin Complex in Lung Adenocarcinoma. Clin Cancer Res. 2017;23:6336–50.
Aakko S, Straume AH, Birkeland EE, Chen P, Qiao X, Lonning PE, Kallio MJ. MYC-induced miR-203b-3p and miR-203a-3p control Bcl-xL expression and paclitaxel sensitivity in tumor cells. Transl Oncol. 2019;12:170–9.
Capizzi M, Strappazzon F, Cianfanelli V, Papaleo E, Cecconi F. MIR7-3HG, a MYC-dependent modulator of cell proliferation, inhibits autophagy by a regulatory loop involving AMBRA1. Autophagy. 2017;13:554–66.
Hong D, Fritz AJ, Gordon JA, Tye CE, Boyd JR, Tracy KM, Frietze SE, Carr FE, Nickerson JA, Van Wijnen AJ, et al. RUNX1-dependent mechanisms in biological control and dysregulation in cancer. J Cell Physiol. 2019;234:8597–609.
Rossetti S, Sacchi N. RUNX1: a microRNA hub in normal and malignant hematopoiesis. Int J Mol Sci. 2013;14:1566–88.
Stavast CJ, Leenen PJM, Erkeland SJ. The interplay between critical transcription factors and microRNAs in the control of normal and malignant myelopoiesis. Cancer Lett. 2018;427:28–37.
Teng H, Wang P, Xue Y, Liu X, Ma J, Cai H, Xi Z, Li Z, Liu Y. Role of HCP5-miR-139-RUNX1 feedback loop in regulating malignant behavior of Glioma cells. Mol Ther. 2016;24:1806–22.
Miao YS, Zhao YY, Zhao LN, Wang P, Liu YH, Ma J, Xue YX. MiR-18a increased the permeability of BTB via RUNX1 mediated down-regulation of ZO-1, occludin and claudin-5. Cell Signal. 2015;27:156–67.
Tang W, Yu F, Yao H, Cui X, Jiao Y, Lin L, Chen J, Yin D, Song E, Liu Q. miR-27a regulates endothelial differentiation of breast cancer stem like cells. Oncogene. 2014;33:2629–38.
Browne G, Dragon JA, Hong D, Messier TL, Gordon JA, Farina NH, Boyd JR, VanOudenhove JJ, Perez AW, Zaidi SK, et al. MicroRNA-378-mediated suppression of Runx1 alleviates the aggressive phenotype of triple-negative MDA-MB-231 human breast cancer cells. Tumour Biol. 2016;37:8825–39.
Acknowledgements
The authors appreciate all the participants and the patients for making contributions to this work.
Funding
This study was supported by Fund of National Natural Science Foundation of China (NSFC81760420, NSFC 81560469), Natural Science Foundation of Guangxi, China (2017GXNSFAA198016, 2016GXNSFAA380255), Guangxi Degree and Postgraduate Education Reform and Development Research Projects, China (JGY2019050), Guangxi Medical University Training Program for Distinguished Young Scholars, Medical Excellence Award Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University, Guangxi Zhuang Autonomous Region Health and Family Planning Commission Self-financed Scientific Research Project (Z20180979).
Author information
Authors and Affiliations
Contributions
CYL and ZYL contributed equally to this work, they finished acquisition and analysis of the data, as well as wrote the manuscript; BLG and WJC participated and performed data analysis in this work; ZBF and GC conceived this project and revised the manuscript. TQG, YYF, YWD and KS collected the samples, performed RT-qPCR and participated in the interpretation of data. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The research proposal was ratified by Committee on Ethics of the First Affiliated Hospital of Guangxi Medical University. Consent was obtained from each subject or their legally authorized representative at the time of enrollment.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liang, CY., Li, ZY., Gan, TQ. et al. Downregulation of hsa-microRNA-204-5p and identification of its potential regulatory network in non-small cell lung cancer: RT-qPCR, bioinformatic- and meta-analyses. Respir Res 21, 60 (2020). https://doi.org/10.1186/s12931-020-1274-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12931-020-1274-9