Abstract
The genome of Helicobacter pylori contains many putative genes, including a genetic region known as the Integrating Conjugative Elements of H. pylori type four secretion system (ICEHptfs). This genetic regions were originally termed as “plasticity zones/regions” due to the great genetic diversity between the original two H. pylori whole genome sequences. Upon analysis of additional genome sequences, the regions were reported to be extremely common within the genome of H. pylori. Moreover, these regions were also considered conserved rather than genetically plastic and were believed to act as mobile genetic elements transferred via conjugation. Although ICEHptfs(s) are highly conserved, these regions display great allele diversity, especially on ICEHptfs4, with three different subtypes: ICEHptfs4a, 4b, and 4c. ICEHptfs were also reported to contain a novel type 4 secretion system (T4SS) with both epidemiological and in vitro infection model studies highlighting that this novel T4SS functions primarily as a virulence factor. However, there is currently no information regarding the structure, the genes responsible for forming the T4SS, and the interaction between this T4SS and other virulence genes. Unlike the cag pathogenicity island (PAI), which contains CagA, a gene found to be essential for H. pylori virulence, these novel T4SSs have not yet been reported to contain genes that contribute significant effects to the entire system. This notion prompted the hypothesis that these novel T4SSs may have different mechanisms involving cag PAI.
Similar content being viewed by others
Background
Helicobacter pylori is one of the most successful pathogenic bacteria that colonizes the human stomach, an organ that had previously been considered to be sterile. Colonization of the human stomach resulted in an evolutionary pressure that prompted H. pylori to acquire genetic adaptations, leading to high genetic diversity in its genome. The high genetic diversity of bacteria can be attributed to many mechanisms, such as genetic drift, horizontal gene transfer, mutations, and migrations [1]. The rapid evolution process that occurs within the genome of H. pylori affects many putative genes that can be divided into three groups: the first group contains genes with variable structures/genotypes depending on the strain. The most well studied example within this group is CagA, which contains a C-terminal repeat segment with a Glu-Pro-Ile-Tyr-Ala (EPIYA) motif and its surrounding region, comprises the EPIYA segment, known as EPIYA-A, -B, and -C/−D, and can discriminate Western-type CagA (with EPIYA-C) and East-Asian-type CagA (with EPIYA-D) [2]. The second group contains the phase-variable genes, the status of which can be altered during the colonization process or in different environments [3]. The best example of genes in this group is the outer membrane protein family. One example is of the blood group antigen binding adhesin (BabA) that is negatively selected during the infection process in the animal models: Rhesus macaques, Mongolian gerbils, and mice [4,5,6]. Another example is the slipped-strand mispairing mechanism, which can change the functional status of the gene, as is the case in the CT repeat of the outer inflammatory protein (OipA) [7]. The last group contains strain-specific genes, including the cag pathogenicity island (PAI), which is among the most well studied and has been reviewed extensively by Backert et al. [8].
In addition to the cag PAI, another intriguing putative gene cluster is the Integrating Conjugative Elements (ICEs) of H. pylori of type four secretion system (ICEHptfs), which has not been studied as much as other virulence factors such as cag PAI and VacA. The fact that this gene cluster has been overlooked can be attributed to inconsistencies and confusion regarding the definition of ICEHptfs. However, with the development of next-generation sequencing (NGS) technology and bioinformatics tools in recent years, we have been able to simplify the definition of ICEHptfs. Besides the more consistent definition of ICEHptfs, current findings highlight the heterogeneity of ICEHptfs, which can be subdivided into ICEHptfs4a/4b/4c and ICEHptfs3 [9]. Furthermore, the association of this cluster with the clinical outcomes of infected patients has been reported. In this review, we focus on the current understanding of the ICEs of H. pylori in terms of global distribution, heterogeneity, and their association with clinical outcomes.
Integrating conjugative elements of H. pylori
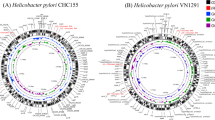
Initially ICEHptfs were defined as the plasticity zones or plasticity regions, as they represent potions of the H. pylori genome with a considerably lower G + C content (approximately 35%) than the rest of the genome (approximately 39%) (Fig. 1) [10]. The low G + C ratio within these regions were considered a result of horizontal gene transfer from an outside source. Another region within the H. pylori genome with low G + C content is cag PAI (Fig. 1). However, due to the conserved regions of cag PAI between the first 2 fully-sequenced strains, J99 [11] and 26,695 [12], these regions were not considered as plasticity zones. Other low G + C content regions differed between J99 and 26,695; therefore, to address the great diversity between the two strains, these regions became known as plasticity zones [3].
Identification of ICEHptfs based on the G + C content from strain 26,695 (Accession: AE000511.1). The G + C ratio was calculated using EMBOSS [58] of each ORFs. The graph was generated based on the average G + C ratio of 10 ORFs. In the strain 26,695 genome, there are three locations that had significantly low G + C ratio, two of which are ICEHptfs and one is cag PAI
Several years later, the plasticity zones were classified as mobile genetic elements (MGEs) [13]. An MGE is a type of genetic material that can move around within the genome and can be transferred from one species or replicon to another. MGEs can be found in every organism, including bacteria, archaea, and humans [14]. MGEs can have different roles in organism evolution because of the various acquisition sources mediated by HGT. In addition, gene duplication of various components within MGEs has been reported. The MGE can be divided into different types, including transposons, plasmids, and bacteriophages [14]. Transposons are DNA sequences that can move within the genome and include both retrotransposons and DNA transposons. The fundamental difference between retrotransposons and DNA transposons is the requirement of an RNA intermediate in the retrotransposon. Plasmids are a collection of functional genetic modules that are organized into stable, self-replicating entities or “replicons,” which are smaller than the cellular chromosome and usually do not contain any essential functional genes. Bacteriophage refers to a virus that replicates within the chromosome of a bacteria [14].
In the case of plasticity zones within H. pylori, the MGEs in the plasticity zones have similar properties to the transposons, and therefore became known as transposon of plasticity zones (TnPZ) (Table 1) [13]. Kersulyte et al. revealed 7 open reading frames (ORFs) of this 16-kb MGE, which are homologues of the VirB system from Agrobacterium tumefaciens, including VirB4, VirB7, VirB8, VirB9, VirB10, VirB11, and VirD4, and therefore is considered as a novel type 4 secretion system (T4SS), termed T4SS3 [15]. This cluster was originally identified in the H. pylori strain PeCan18B, which was isolated from a patient with gastric cancer in Peru. Interestingly, subsequent studies investigating additional strains from different locations worldwide using polymerase chain reaction (PCR)-Sanger sequence method [13] and NGS [9, 16] showed these MGEs, which were originally thought as “plasticity zones” with high genomic variability [10, 11], were in fact highly conserved. These MGEs were also predicted to have been transferred into the H. pylori genome via conjugation as ICEs and are known as ICEHptfs in H. pylori [9]. The ICEs on many other bacteria were typically transferred via the following mechanism [17]. First, the element is typically excised from the original chromosome by the recombinase to create circular intermediate. The circular intermediate is then transferred from the donor to the recipient cell via conjugation. Lastly, the ICEs integrate into the recipient cell chromosome via site-specific or non-specific recombination. In the case of ICEHptfs4, the excision step is dependent on XerT recombinase [16], and the conjugation process was dependent on the VirD2 relaxase [18]. The motif for the integration site-specific recombination is most likely AAGAATG (or possibly AAGAAT for ICEHptfs3) [9]. Given the low specificity of ICEs, there are over 100,000 possible integration sites. However, only 40 different integration site motifs have been reported in the case of H. pylori. Given that the AAGAATG integration site motif was identified at 550 sites within the H. pylori genome, the possible integration sites might be higher [9]. In this regard, the integration site of ICEs in H. pylori was considered to possess intermediate specificity.
Based on the newest findings regarding the H. pylori MGE, the most recent proposed name of these particular mobile elements is ICEHptfs. In this review, we will refer to these MGE(s), including the TFSS inside of these elements, as ICEHptfs and we will mention the original name from the cited paper. The cag PAI indeed had a similar features to ICEHptfs, such as low G + C content, and was flanked by a 31 bp motif representing the integration site [19] and contained the VirB homologue forming cag T4SS to translocate CagA [8, 20]. However, upon comparison of the 29 H. pylori genomes, the cag PAI was classified as a core genome instead of mobile gene clusters, as is the case for ICEHptfs3 or ICEHptfs4 [21].
The genetic diversity of ICEHptfs among H. pylori strains worldwide
The study of the prevalence of ICEHptfs was initiated in the strains from Costa Rica, isolated from patients with gastritis and gastric cancer in 2000 [22]. Using the dot blot method to determine the presence or absence of ICEHptfs genes in strain J99 (known as the “plasticity region” genes), variation among the Costa Rican strains ranged from 17% (jhp0940) to 100% (jhp0912) [22]. Subsequent studies used a H. pylori genome microarray approach to detect the presence or absence of strain J99 ICEHptfs genes [23, 24]. Those studies investigated a total of 56 [23] and 15 strains [24] worldwide. The prevalence of the J99 ICEHptfs from those 71 strains showed that the prevalence of ORFs in the J99 ICEHptfs varied among the studied strains, except the jhp0915, which were present in all studied strains [3]. Although the original purpose of detecting J99 ICEHptfs genes was to screen the candidate genes for genetic markers of clinical outcomes, there were no significant findings until the discovery of a new cluster of T4SS on ICEHptfs. In addition, a subsequent study to investigate the distribution of ICEHptfs using a PCR-based method with 16 representative ICEHptfs ORFs (J99 [11 ORFs], 26,695 [3 ORFs], PeCan18B [1 ORF], and CPY6081 [1 ORF]) to 102 strains variously isolated from Spain, Japan, India, Peru, and Gambia reported ORFs in almost all (92.15%, 94/102) screened strains, with an average of 6 ORFs per strain [13]. The most commonly found ICEHptfs on the H. pylori were also reported upon whole genome analysis of 45 [9] and 218 [25] strains of H. pylori, for which ICEHptfs were detected in 31/45 (68.8%) and 204/218 (93.5%) strains, respectively. However, our recent study in Indonesia reported the prevalence of ICEHptfs was lower than that in previous reports and ICEHptfs were reported in only 54.3% (56/103) of the analyzed strains [26]. In addition, ICEHptfs were also absent in several CagA types, such as ABBD, AABD, ABCC, and B type CagA. The absence of ICEHptfs in the exclusive CagA genotypes suggests that the distribution of ICEHptfs might be associated with the population genetic of H. pylori [26].
Aside from the prevalence of ICEHptfs, which could be locus-dependent, there was an interesting finding on the allele of ICEHptfs within the H. pylori genome [9, 13, 16]. The first global analysis of ICEHptfs in 2009 discovered several types of ICEHptfs(s), called as ICEHptfs type 1, type 1band type 2, which also contained a different type of T4SS called T4SS3a, T4SS3b, and T4SS3 (referred to as tfs3, tfs3b, and tfs3 in the study), respectively (Table 1) [13]. In 2010, however, Fischer et al. proposed to change the name of type 1b/1 and type 2 to ICEHptfs4a/4b and ICEHptfs3, respectively (Table 1) [9, 16]. The difference of these genetic elements was determined based on the general structure of ICEHptfs themselves, which showed the location of putative methylase/helicase was directly adjacent to the virD4 homologue on ICEHptfs3 and the parA homologue on ICEHptfs4 [15]. In addition, the type of the T4SS in ICEHptfs could also be distinguished. The analysis of the most conserved region (virB9, virB10, and virD4) of the T4SS in ICEHptfs showed a super lineage between T4SS4 and T4SS3 [9]. In the T4SS4 group, there were three subtypes called T4SS4a, T4SS4b, and T4SS4c (Fig. 2). Both T4SS4a and 4b were most commonly found to contain ICEHptfs4, whereas T4SS4c was only present in the strains from South Africa [9]. As a comparison, the similar genetic cluster occurred because of horizontal gene transfer; the phylogenetic analysis of the cag PAI showed there was a linear correlation between the cag PAI and the population genetic generated by the multi-locus sequence typing (MLST) [27]. However, the study conducted to analyze 218 H. pylori genome sequences showed the there was no correlation in terms of co-occurrence in either the status or type of ICEHptfs with the cag PAI [25], suggesting an association to the H. pylori population genetic, although the evolution pattern may be different than that of the cag PAI.
Genetic properties of ICEHptfs3 and ICEHptfs4 based on the Fischer et al. classifications [9]. The red arrow indicates the T4SS-forming genes. The striped pattern indicates a different sequence compared to ICEHptfs4a. ICEHptfs3 and ICEHptfs4 had different overall genetic structures. ICEHptfs4 had three subtypes based of different alleles of the T4SS-forming genes and other accessories genes
The analysis of ICEHptfs from hundreds of genome sequences has highlighted another way to classify the various types of ICEHptfs. The classification was created based on the conservative-variable analysis of the structure on a left-central-right segment of the ICEHptfs4 region and on the left-right segment of ICEHptfs3. In ICEHptfs4, the classification was as follows: the left segment as the L1/L2/Lm, central segment as C1/C2, and right segment as the R1/R2/R1f (Fig. 3) [25]. Based on the previous classification, which could differentiate between ICEHptfs4a/4b/4c, utilizing current classification it could be determined as L2-C1-R2 for ICEHptfs4a, L1-C1-R1 for ICEHptfs4b, and Lm-C2-R2 for ICEHptfs4c. Analysis of the type of segments to the population genetic based on the MLST showed a ubiquitous presence of ICEHptfs4, especially the L1-C1-R1 subtype, which may be associated with the ancestral population of H. pylori prior to spatial separation [25]. In addition, the C1 subtype was reported to be present in H. acinonychis, which is in the same clade as the super lineage of the hpAfrica2 population [28], supporting the existence of an ancient association of this particular element. In contrast, the L2-C2-R2 subtype module had a lower abundance than the type 1 counterpart, suggesting the presence of a different evolutionary history, which may be associated with the recent acquisition and adaptation to a particular isolate of H. pylori [25].
New classification based on orthologue analysis of 218 ICEHptfs genomes adapted from Delahay et al. [25]. a The modular subtype of ICEHptfs4. The modular subtype was classified based on three regions: left flank (L1/L2/Lm), central region (C1/C2), and right flank (R1/R2/R1f). The green arrow corresponds to the conserved region, whereas the blue and red arrows correspond to the type 1 (L1-C1-R1/R1f) and type 2 (L2-C2-R2) modules, respectively. The Lm was described as a combination between L1 and L2 on the left flank region. b The modular type of ICEHptfs3. Unlike ICEHptfs4, which had a subtype, ICEHptfs3 is conserved in almost ICEHptfs3-containing strains with the classification based on the presence/absence of the left segment and the right segment of this region
In ICEHptfs3, there was no modular subtype of the segments as is the case for ICEHptfs4, which can be differentiated as type 1 and 2. In addition, the distribution of ICEHptfs3 was considerably lower than that of ICEHtpfs4, and this particular type was more commonly found as an incomplete cluster than the ICEHptfs4 [25]. This observation was also agreement with our previous findings highlighting that the degree of complete clustering of ICEHptfs3 was lower than that of ICEHptfs4 [26]. In addition, the left segment of ICEHptfs3 was more frequent than the right segment and had a higher co-occurrence with ICEHptfs4 L modules and/or cag PAI, suggesting potential differences in stability or temporal acquisition of ICEHptfs3 [25]. Notably, this phenomenon was commonly found in the strain from the hspAmerind population [25]. These data suggest that even ICEHptfs, which were considered to have different evolutionary history than other mobile-like elements like cag PAI, still have a characteristic of particular clades of H. pylori population, which showed a tendency to play a role as a donor than a recipient [25, 29].
The role of ICEHptfs in H. pylori infection
The MGEs are always transferred to a particular genome with a purpose. One of the reasons behind the transferring process is a fitness advantage, including increased virulence. The most well studied system to confer virulence in H. pylori is the T4SS. In fact, the T4SS is found in all known ICEs, genomic islands, and conjugative plasmids of Gram-negative bacteria, except those of Bacteroides species [14]. In the case of H. pylori, ICEHptfs were predicted to have a genetic advantage as clinical outcome predictors. Indeed, at the initial discovery of this MGE, most of the gene members were hypothetical. However, several genes were predicted to be homologues of the A. tumefaciens VirB T4SS components, with other genes being involved in the horizontal gene transfer process, such as XerT, TopA, and ParA [15]. Therefore, most of the studies related to this MGE highlight the association of the genes within these elements to clinical outcome.
The first study was conducted to screen the candidate genes of strain J99 ICEHptfs (“plasticity region” in the study) in the Costa Rican patients, resulting in several genes such as jhp0940 and jhp0947 as predictors for gastric cancer development and hp0986, which was associated with the prevalence of gastritis [22]. The subsequent study involving 200 patients from Brazil with variable clinical backgrounds, including gastric cancer, duodenal ulcer, and gastritis, confirmed that jhp0947 was related to the development of gastric cancer with an odds ratio (OR) of 4.14 (95% confidence interval [CI] = 1.47–11.66) [30]. In addition, jhp0947 was found to frequently co-exist with jhp0949 [31, 32]. Together, these two genes were associated with the induction of interleukin-12 (IL-12) and a higher prevalence of duodenal ulcer [31]. In addition to the induction of IL-12, the jhp0947 together with jhp0945 and jhp0949 induced significantly higher IL-8 and faster apoptosis in the cell lines [33]. The jhp0947 gene was found to be homologous to jhp0938 (hp0990) and jhp0253 (hp1333), which all encode hypothetical proteins. However, bioinformatics analysis showed the 5′ region of jhp0947 was also homologous to jhp0477 (hp0528), which is a part of the cag PAI (virB9 homologue) and had been identified as an important structural component of the cag T4SS. Originally, VirB9 from A. tumefaciens contributed to substrate selection and translocation, establishment of channel subunit contacts, and T-pili biogenesis [34]. However, in the case of H. pylori homologue in the T4SS4 and/or T4SS3, the function of this gene is yet to be described. Therefore, further studies to elucidate the function are necessary.
In addition, the VirB4 homologue (jhp0917/jhp0918) of ICEHptfs4b, duodenal ulcer promoting (DupA) was predicted to be important in the development of duodenal ulcer and prevention of gastric carcinoma [35]. dupA was screened together with other virB homologues within the H. pylori genome and was found to be truncated into two genes in the strain J99. The truncated gene was a result of a 1 bp insertion after the position 1385 in strain C145 (GenBank Accession number AB196363), which resulted in a frameshift mutation. However, strain J99 was a rare example in which dupA was truncated into two genes. As expected, the screening of jhp0917 and jhp0918 revealed that they were always co-existed [35], suggesting that virB4 is a continuous gene of both jhp0917 and jhp0918. A subsequent study in Brazilian strains showed that among 89 strains studied, 86 strains (97%) contained a 1 bp insertion at position 1385 [36], confirming the hypothesis that intact virB4 did not contain a 1 bp insertion downstream of position 1385.
Functional prediction analysis showed that DupA was predicted to be the homologue to virB4 ATPase, which is involved in the DNA uptake/DNA transfer and protein transfer of MGEs. The 5′ terminus of the dupA, which is encoded by the jhp0917, specifically on the location 3–201, has homology to the FtsK/SpoIIIE family of ATPases [35]. FtsK/SpoIIIE family ATPases are conserved throughout bacteria and are involved in the translocation of DNA and proteins through membrane-spanning pores [37]. In addition, proteins within this family contain a putative ATP-binding P-loop motif, are involved in cell division and peptidoglycan synthesis/modification, and have been implicated in intercellular chromosomal DNA transfer. The central region of dupA, which is encoded by jhp0917 5′–jhp0918, specifically at position 203–610, showed homology to the TraG/TraD family protein [35]. TraG-like proteins are potential NTP hydrolases (NTPases) that are essential for DNA transfer in bacterial conjugation and may mediate the interaction between DNA processing and mating formation systems [38].
The importance of cluster genes rather than a single gene
The association between the pathogen and the infected host involves a complex interaction between the bacterial genetics, host vulnerability, and the environment of the infection. With respect to bacterial genetics, most of the reported findings highlight that a single gene or cluster of genes is responsible for the development of severe clinical outcomes of the infected individual. In H. pylori, the most well studied virulence gene is cagA, which encodes the oncogenic protein CagA. CagA is a member of cag PAI, a T4SS complex that can form a pilus to surround H. pylori, allowing for the transfer of the CagA protein into the host cell [8]. There are many studies showing the association of cagA to the prevalence of gastric carcinoma in the Western population [39,40,41]. However, nearly all of the isolated strains from East-Asian countries contained cagA, and therefore the presence or absence of CagA alone does not discriminate the clinical outcomes of infected individuals [2]. Hence, it was proposed that the heterogeneity of the C-terminus of CagA, which includes repeat segment containing EPIYA motif and its surrounding region, comprises the EPIYA segment, known as EPIYA-A,-B, and -C/−D, and can discriminate between Western-type CagA and East-Asian-type CagA, respectively [2]. East-Asian-type CagA has a higher affinity for the SH2 domain, which can result in a worse cascade output directly following the initial infection [42]. However, our recent study exploring the correlation of CagA heterogeneity status with the clinical outcome did not show a promising result [43,44,45]. Our observation in Indonesia showed that individuals infected with East-Asian-type CagA strains had even lower inflammation scores than those infected with Western-type CagA strains [44]. In addition, reports from two locations in Thailand with different gastric cancer incidences further contradicted previous findings, as individuals infected with Western-type CagA strains were found to have significantly higher antral activity than those with the East-Asian-type CagA strains [45]. Moreover, our observation in Mongolia, the second highest gastric carcinoma prevalence based on age standardization rate (ASR); GLOBOCAN, 2012 (http://globocan.iarc.fr) (ASR = 47.4/100,000 men), showed our developed East-Asian specific CagA antibody [46] had negative results in most of immunohistochemistry biopsy specimens [43], suggesting the most CagA type in Mongolia was Western type. These data suggest that the CagA is still essential, but in certain areas, a complex interaction between infected individuals and the pathogen to generate a clinical outcome may not be attributed to a single gene. Therefore, it is interesting to investigate additional genes in close proximity that may be involved (e.g. cagA and cag PAI, dupA cluster) or genes from another system that may interact in an epistatic manner.
Initial observations of cag PAI intactness to allele diversity of vacA showed the intact contiguous cag PAI was frequently found in patients with peptic ulcer diseases (78%) and gastric carcinoma (73%) and that this frequency was significantly higher than that in the gastritis patients (40%, p < 0.01) [47]. Among the virulence factors, there was an association between intact cag PAI and both the vacA s1 allele and m1 allele (p < 0.005 and p = 0.05, respectively) [47]. A subsequent study in Sweden showed that H. pylori isolates containing all the genes within the cag PAI induced high IL-8 production in AGS cells, a gastric cancer cell line. Additionally, the presence of intact cag PAI was reported to have more than five-times higher risk of developing more severe gastroduodenal diseases than the absence of cag PAI (95% CI = 1.5–17.4) [48]. Another observation of the association between intact cag PAI to the clinical outcome was also reported in Iran [49]. The patients infected with intact cag PAI strains were reported to have more severe atrophy than those with non-intact cag PAI strains [49]. Our observation in Vietnam similarly showed that there was an association of intact cag PAI H. pylori with patients with peptic ulcers, as all of the enrolled peptic ulcer patients were infected with intact cag PAI H. pylori [50]. In addition, patients infected with intact cag PAI had higher inflammation scores in the antrum, corpus, and upper lesser curvature [50]. In our recent study, we observed that in Indonesia, the cag PAI was identified in almost all Indonesian H. pylori isolates (99%), but was associated with various clinical outcomes. However, after detailed analysis of the gene functionality, we found that several genes contained either a premature stop codon or a frameshift mutation within the cag PAI genes. Therefore, we consider a region containing a non-functional gene to represent non-intact cag PAI. These findings suggest that the intactness of cag PAI does not solely depend on the presence or absence of the cag PAI-forming genes and also depends on the functional status of the forming genes [26]. These findings suggest that the importance of virulence factors may be determined by a cluster of related genes, rather than a single gene in isolation.
The concept of a cluster of the genes, which may more accurately predict the clinical outcome of the infected individual, is also relevant to ICEHptfs genes. In 2005, our group discovered dupA, which had a strong positive correlation with the prevalence of duodenal ulcer, but a negative correlation with gastritis atrophy, intestinal metaplasia, and gastric cancer [35]. Indeed, the subsequent systematic review and meta-analysis involving 17 studies with 2466 patients of dupA showed a positive correlation between dupA and the prevalence of duodenal ulcer in the general population (OR = 1.41, 95% CI = 1.12–1.76), while subsequent analysis in the Asian population showed an OR = 1.57 (95% CI = 1.19–2.06) [51]. However, in the Western population, dupA did not show any correlation with the prevalence of duodenal ulcer (OR = 1.09, 95% CI = 0.73–1.62) [51].
Notably, there were several inconsistent results in studies that followed the discovery of dupA. dupA, a homologue of VirB4 from ICEHptfs4b [9], was reported to have an extra 600 bp on the 5′ end of the original dupA gene (jhp0917-jhp0918). The variant was referred to as dupA long type [52], with two different functional status characterized as long intact and long non-intact depending on the presence/absence of a 1 bp insertion at position 1385 in strain C145, resulting in a frameshift mutation [35]. In the Okinawan population in Japan, long-intact dupA significantly increased the risk of infected individuals to develop gastric cancer and ulcers rather than gastritis by more than 3-fold and 4-fold (OR = 3.3, 95% CI = 1.55–7.24 and OR = 4.14, 95% CI = 1.23–13.94), respectively [52]. In addition to the observation of a 1 bp insertion, the dupA was reported to have another allele polymorphism with an adenine deletion at position 1131 and an adenine insertion at position 1426. These insertions and deletions can lead to premature stop codons, which may produce a non-functional gene. We observed that this allele polymorphism leading to intact dupA (an allele without any insertions and deletions leading to a premature stop codon) was associated with the increase in mucosal inflammation, but an overall decrease in mucosal atrophy. Moreover, the intact dupA was negatively associated with gastric carcinoma [53]. These findings suggest that the long-intact dupA was more reliable as a clinical outcome predictor than the short type dupA.
Notably, dupA is surrounded by several genes such as virB8, virB9, virB10, and virB11 which can form the T4SS, named as T4SS4b, which lies within ICEHptfs4b. Therefore, once all the components necessary for forming the T4SS have been identified, we will gain a better understanding of using this biomarker to predict the clinical outcome of infected patients. Our epidemiological observation of the dupA and its cluster forming the T4SS showed that the complete dupA cluster was significantly correlated with the prevalence of duodenal ulcer in the US population [54]. In addition, individuals infected with H. pylori containing the intact dupA cluster showed a higher atrophy score in both the antrum and corpus. In the in vitro model, isolates with intact dupA cluster H. pylori could induce significantly higher IL-8 production in both gastric epithelial cells and the gastric cancer cell line MKN45 [54]. Our latest observation in Indonesia showed the intact ICEHptfs4b was the most associated with increased inflammation in the antrum compared with ICEHptfs-negative status [26]. In addition, in combination with the cag PAI, patients infected with H. pylori containing both intact cag PAI and ICEHptfs4b had the highest inflammation, both in the antrum and corpus [26]. Furthermore, in H. pylori isolates from pediatric patients, the virB4-like gene dupA was not associated with any differences in IL-8 production and NF-κB phosphorylation in infected gastric cells. However, the complete cluster of dupA H. pylori isolates, which encoded T4SS4b, induced significantly higher IL-8 production in infected gastric epithelial cell lines [55]. These findings when taken together suggest several points. Firstly, dupA alone is an essential factor for determining the clinical outcome of an infected individual. This has also been shown using in vitro studies [35], although there are several discrepancies between different populations. Second, dupA and its other homologues from ICEHptfs4a/3 play a role in the formation of the T4SS, and are therefore more predictive of disease than a single gene.
In addition to the role of dupA, which belongs to ICEHptfs4b, a recent study had reported a role of ICEHptfs3 in the H. pylori infection process. One of the ICEHptfs forming genes was predicted to encode cell translocating kinase A (CtkA). CtkA is a protein that induces a pro-inflammatory response within infected host cells [56]. Despite the previous understanding that CtkA was considerably more variable in different geographic populations, a recent study showed that CtkA was encoded by one of the genes within ICEHptfs3, which are located closer to xer and virD2 toward the end of the ICE in H. pylori and H. cetorum [57]. In addition, CtkA-induced expression of pro-inflammatory cytokines was dependent on ICEHptfs3, but independent of the cag PAI. The induction of pro-inflammatory cytokines within the infected cell lines was mediated by the activation of NF-κB [57]. These data confirm the importance of ICEHptfs3 during H. pylori infection, in particular through CtkA as one of the substrates of this genetic island.
Conclusion
Since the discovery of this particular gene region in the H. pylori genome, there have been a variety of proposed names including the plasticity region, plasticity zones, TnPZ, and ICEHptfs, which has led to the confusion regarding the terminology. Based on new findings regarding this gene cluster, the most relevant name is ICEHptfs. With the increasing number of available genome sequences, it has been reported that this region is commonly present in H. pylori genome with some variability in the different geographic areas. In addition, ICEHptfs were also reported to have novel T4SS(s) with functions related to virulence. However, the function of each component forming the full set as well as the structure of this novel T4SS are yet to be described. Therefore, future studies to elucidate the genetic components of the T4SS, the structure of the formed T4SS, and the role of each gene in T4SS function are necessary. Unlike the cag PAI, which has the CagA gene that has been shown to be essential for virulence, no genes with significant effects on the entire system have been identified within these novel T4SSs. This condition leads to the hypothesis that these novel T4SSs may have different mechanism involving the cag PAI.
Abbreviations
- ASR:
-
Age standardization rate
- CI:
-
Confidence interval
- CtkA:
-
Cell translocating kinase A
- ICE:
-
Integrating conjugative element
- IL:
-
Interleukin
- MGE:
-
Mobile genetic element
- MLST:
-
Multi locus sequence typing
- NGS:
-
Next generation sequencing
- OR:
-
Odd ratio
- ORF:
-
Open reading frame
- PAI:
-
Pathogenicity island
- PCR:
-
Polymerase chain reaction
- T4SS:
-
Type IV secretion system
- TnPZ:
-
Transposon of plasticity zones
References
Weale ME. Book review: balding DJ, bishop M, Cannings C 2007: handbook of statistical genetics, 3rd edition, Chichester: Wiley, 1616 p. (HB). ISBN: 978-0-470-05830-5, £195. Stat Methods Med Res. 2010;19(2):197.
Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7(11):629–41.
Yamaoka Y. Roles of the plasticity regions of Helicobacter pylori in gastroduodenal pathogenesis. J Med Microbiol. 2008;57(Pt 5):545–53.
Kable ME, Hansen LM, Styer CM, Deck SL, Rakhimova O, Shevtsova A, Eaton KA, Martin ME, Gideonsson P, Boren T, Solnick JV. Host determinants of expression of the Helicobacter pylori BabA Adhesin. Sci Rep. 2017;7:46499.
Ohno T, Vallstrom A, Rugge M, Ota H, Graham DY, Arnqvist A, Yamaoka Y. Effects of blood group antigen-binding adhesin expression during Helicobacter pylori infection of Mongolian gerbils. J Infect Dis. 2011;203(5):726–35.
Solnick JV, Hansen LM, Salama NR, Boonjakuakul JK, Syvanen M. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc Natl Acad Sci U S A. 2004;101(7):2106–11.
Yamaoka Y, Kikuchi S, el-Zimaity HM, Gutierrez O, Osato MS, Graham DY. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology. 2002;123(2):414–24.
Backert S, Haas R, Gerhard M, Naumann M. The Helicobacter pylori type IV secretion system encoded by the cag pathogenicity island: architecture, function, and signaling. Curr Top Microbiol Immunol. 2017;413:187–220.
Fischer W, Breithaupt U, Kern B, Smith SI, Spicher C, Haas R. A comprehensive analysis of Helicobacter pylori plasticity zones reveals that they are integrating conjugative elements with intermediate integration specificity. BMC Genomics. 2014;15:310.
Alm RA, Trust TJ. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J Mol Med. 1999;77(12):834–46.
Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397(6715):176–80.
Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388(6642):539–47.
Kersulyte D, Lee W, Subramaniam D, Anant S, Herrera P, Cabrera L, Balqui J, Barabas O, Kalia A, Gilman RH, Berg DE. Helicobacter pylori’s plasticity zones are novel transposable elements. PLoS One. 2009;4(9):e6859.
Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3(9):722–32.
Kersulyte D, Velapatino B, Mukhopadhyay AK, Cahuayme L, Bussalleu A, Combe J, Gilman RH, Berg DE. Cluster of type IV secretion genes in Helicobacter pylori’s plasticity zone. J Bacteriol. 2003;185(13):3764–72.
Fischer W, Windhager L, Rohrer S, Zeiller M, Karnholz A, Hoffmann R, Zimmer R, Haas R. Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 2010;38(18):6089–101.
Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8(8):552–63.
Grove JI, Alandiyjany MN, Delahay RM. Site-specific relaxase activity of a VirD2-like protein encoded within the tfs4 genomic island of Helicobacter pylori. J Biol Chem. 2013;288(37):26385–96.
Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R. Covacci a. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A. 1996;93(25):14648–53.
Backert S, Tegtmeyer N, Fischer W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol. 2015;10(6):955–65.
Uchiyama I, Albritton J, Fukuyo M, Kojima KK, Yahara K, Kobayashi I. A novel approach to Helicobacter pylori pan-genome analysis for identification of genomic islands. PLoS One. 2016;11(8):e0159419.
Occhialini A, Marais A, Alm R, Garcia F, Sierra R, Megraud F. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect Immun. 2000;68(11):6240–9.
Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, Yamaoka Y, Kraft C, Suerbaum S, Meyer TF, Achtman M. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1(4):e43.
Salama N, Guillemin K, McDaniel TK, Sherlock G, Tompkins L, Falkow S. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc Natl Acad Sci U S A. 2000;97(26):14668–73.
Delahay RM, Croxall NJ, Stephens AD. Phylogeographic diversity and mosaicism of the Helicobacter pylori tfs integrative and conjugative elements. Mob DNA. 2018;9:5.
Waskito LA, Miftahussurur M, Lusida MI, Syam AF, Suzuki R, Subsomwong P, Uchida T, Hamdan M, Nasronudin, Yamaoka Y. Distribution and clinical associations of integrating conjugative elements and cag pathogenicity islands of Helicobacter pylori in Indonesia. Sci Rep. 2018;8(1):6073.
Olbermann P, Josenhans C, Moodley Y, Uhr M, Stamer C, Vauterin M, Suerbaum S, Achtman M, Linz B. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6(8):e1001069.
Moodley Y, Linz B, Bond RP, Nieuwoudt M, Soodyall H, Schlebusch CM, Bernhoft S, Hale J, Suerbaum S, Mugisha L, van der Merwe SW, Achtman M. Age of the association between Helicobacter pylori and man. PLoS Pathog. 2012;8(5):e1002693.
Yahara K, Furuta Y, Oshima K, Yoshida M, Azuma T, Hattori M, Uchiyama I, Kobayashi I. Chromosome painting in silico in a bacterial species reveals fine population structure. Mol Biol Evol. 2013;30(6):1454–64.
Santos A, Queiroz DM, Menard A, Marais A, Rocha GA, Oliveira CA, Nogueira AM, Uzeda M, Megraud F. New pathogenicity marker found in the plasticity region of the Helicobacter pylori genome. J Clin Microbiol. 2003;41(4):1651–5.
de Jonge R, Kuipers EJ, Langeveld SC, Loffeld RJ, Stoof J, van Vliet AH, Kusters JG. The Helicobacter pylori plasticity region locus jhp0947-jhp0949 is associated with duodenal ulcer disease and interleukin-12 production in monocyte cells.FEMS Immunol Med Microbiol. 2004;41(2):161–7.
Sugimoto M, Watada M, Jung SW, Graham DY, Yamaoka Y. Role of Helicobacter pylori plasticity region genes in development of gastroduodenal diseases. J Clin Microbiol. 2012;50(2):441–8.
Ganguly M, Sarkar S, Ghosh P, Sarkar A, Alam J, Karmakar BC, De R, Saha DR, Mukhopadhyay AK. Helicobacter pylori plasticity region genes are associated with the gastroduodenal diseases manifestation in India. Gut pathogens. 2016;8:10.
Jakubowski SJ, Cascales E, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J Bacteriol. 2005;187(10):3486–95.
Lu H, Hsu PI, Graham DY, Yamaoka Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology. 2005;128(4):833–48.
Gomes LI, Rocha GA, Rocha AM, Soares TF, Oliveira CA, Bittencourt PF, Queiroz DM. Lack of association between Helicobacter pylori infection with dupA-positive strains and gastroduodenal diseases in Brazilian patients. Int J Med Microbiol : IJMM. 2008;298(3–4):223–30.
Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32(17):5260–79.
Schroder G, Krause S, Zechner EL, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol. 2002;184(10):2767–79.
Crabtree JE, Wyatt JI, Sobala GM, Miller G, Tompkins DS, Primrose JN, Morgan AG. Systemic and mucosal humoral responses to Helicobacter pylori in gastric cancer. Gut. 1993;34(10):1339–43.
Nomura AM, Lee J, Stemmermann GN, Nomura RY, Perez-Perez GI, Blaser MJ. Helicobacter pylori CagA seropositivity and gastric carcinoma risk in a Japanese American population. J Infect Dis. 2002;186(8):1138–44.
Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40(3):297–301.
Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4(9):688–94.
Matsuhisa T, Yamaoka Y, Uchida T, Duger D, Adiyasuren B, Khasag O, Tegshee T, Tsogt-Ochir B. Gastric mucosa in Mongolian and Japanese patients with gastric cancer and Helicobacter pylori infection. World J Gastroenterol. 2015;21(27):8408–17.
Miftahussurur M, Syam AF, Makmun D, Nusi IA, Zein LH, Zulkhairi, Akil F, Uswan WB, Simanjuntak D, Uchida T, Adi P, Utari AP, Rezkitha YA, Subsomwong P, Nasronudin, Yamaoka Y. Helicobacter pylori virulence genes in the five largest islands of Indonesia. Gut pathogens. 2015;7:26.
Subsomwong P, Miftahussurur M, Uchida T, Vilaichone RK, Ratanachu-Ek T, Mahachai V, Yamaoka Y. Prevalence, risk factors, and virulence genes of Helicobacter pylori among dyspeptic patients in two different gastric cancer risk regions of Thailand. PLoS One. 2017;12(10):e0187113.
Uchida T, Kanada R, Tsukamoto Y, Hijiya N, Matsuura K, Yano S, Yokoyama S, Kishida T, Kodama M, Murakami K, Fujioka T, Moriyama M. Immunohistochemical diagnosis of the cagA-gene genotype of Helicobacter pylori with anti-east Asian CagA-specific antibody. Cancer Sci. 2007;98(4):521–8.
Kidd M, Lastovica AJ, Atherton JC, Louw JA. Conservation of the cag pathogenicity island is associated with vacA alleles and gastroduodenal disease in south African Helicobacter pylori isolates. Gut. 2001;49(1):11–7.
Nilsson C, Sillen A, Eriksson L, Strand ML, Enroth H, Normark S, Falk P, Engstrand L. Correlation between cag pathogenicity island composition and Helicobacter pylori-associated gastroduodenal disease. Infect Immun. 2003;71(11):6573–81.
Ahmadzadeh A, Ghalehnoei H, Farzi N, Yadegar A, Alebouyeh M, Aghdaei HA, Molaei M, Zali MR, Pour Hossein Gholi MA. Association of CagPAI integrity with severeness of Helicobacter pylori infection in patients with gastritis. Pathol Biol (Paris). 2015;63(6):252–7.
Nguyen LT, Uchida T, Tsukamoto Y, Trinh TD, Ta L, Mai HB, Le HS, Ho DQ, Hoang HH, Matsuhisa T, Okimoto T, Kodama M, Murakami K, Fujioka T, Yamaoka Y, Moriyama M. Clinical relevance of cagPAI intactness in Helicobacter pylori isolates from Vietnam. Eur J Clin Microbiol Infect Dis : official publication of the European Society of Clinical Microbiology. 2010;29(6):651–60.
Shiota S, Matsunari O, Watada M, Hanada K, Yamaoka Y. Systematic review and meta-analysis: the relationship between the Helicobacter pylori dupA gene and clinical outcomes. Gut pathogens. 2010;2(1):13.
Takahashi A, Shiota S, Matsunari O, Watada M, Suzuki R, Nakachi S, Kinjo N, Kinjo F, Yamaoka Y. Intact long-type dupA as a marker for gastroduodenal diseases in Okinawan subpopulation, Japan. Helicobacter. 2013;18(1):66–72.
Queiroz DM, Rocha GA, Rocha AM, Moura SB, Saraiva IE, Gomes LI, Soares TF, Melo FF, Cabral MM, Oliveira CA. dupA polymorphisms and risk of Helicobacter pylori-associated diseases. Int J Med Microbiol : IJMM. 2011;301(3):225–8.
Jung SW, Sugimoto M, Shiota S, Graham DY, Yamaoka Y. The intact dupA cluster is a more reliable Helicobacter pylori virulence marker than dupA alone. Infect Immun. 2012;80(1):381–7.
Silva B, Nunes A, Vale FF, Rocha R, Gomes JP, Dias R, Oleastro M. The expression of Helicobacter pylori tfs plasticity zone cluster is regulated by pH and adherence, and its composition is associated with differential gastric IL-8 secretion. Helicobacter. 2017;22(4):8408-17.
Rizwan M, Alvi A, Ahmed N. Novel protein antigen (JHP940) from the genomic plasticity region of Helicobacter pylori induces tumor necrosis factor alpha and interleukin-8 secretion by human macrophages. J Bacteriol. 2008;190(3):1146–51.
Alandiyjany MN, Croxall NJ, Grove JI, Delahay RM. A role for the tfs3 ICE-encoded type IV secretion system in pro-inflammatory signalling by the Helicobacter pylori Ser/Thr kinase, CtkA. PLoS One. 2017;12(7):e0182144.
Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends in genetics : TIG. 2000;16(6):276–7.
Acknowledgements
None.
Funding
This work was supported in part by grants from the National Institutes of Health (DK62813) and the Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (221S0002, 16H06279, 15H02657 and 16H05191) (YY). It was also supported by the Japan Society for the Promotion of Science (JSPS) Institutional Program for Core-to-Core Program; B. Africa-Asia Science Platform (YY). LAW is a doctoral student supported by the Japanese Government (MEXT) Scholarship Program for 2015.
Availability of data and materials
Not applicable.
Author information
Authors and Affiliations
Contributions
YY initiated the idea, guided the article structure, and improved the final manuscript. LAW and JYW reviewed the published studies and composed the draft of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Waskito, L.A., Yih-Wu, J. & Yamaoka, Y. The role of integrating conjugative elements in Helicobacter pylori: a review. J Biomed Sci 25, 86 (2018). https://doi.org/10.1186/s12929-018-0489-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12929-018-0489-2