Abstract
Background
Intestinal tumorigenesis is promoted by myeloid differentiation primary response gene 88 (MyD88) activation in response to the components of microbiota in Apc Min/+ mice. Microbiota also contains double-stranded RNA (dsRNA), a ligand for TLR3, which activates the toll-like receptor adaptor molecule 1 (TICAM-1, also known as TRIF) pathway.
Methods
We established Apc Min/+ Ticam1 −/− mice and their survival was compared to survival of Apc Min/+ Myd88 −/− and wild-type (WT) mice. The properties of polyps were investigated using immunofluorescence staining and RT-PCR analysis.
Results
We demonstrate that TICAM-1 is essential for suppression of polyp formation in Apc Min/+ mice. TICAM-1 knockout resulted in shorter survival of mice compared to WT mice or mice with knockout of MyD88 in the Apc Min/+ background. Polyps were more frequently formed in the distal intestine of Apc Min/+ Ticam1 −/− mice than in Apc Min/+ mice. Infiltration of immune cells such as CD11b+ and CD8α+ cells into the polyps was detected histologically. CD11b and CD8α mRNAs were increased in polyps of Apc Min/+ Ticam1 −/− mice compared to Apc Min/+ mice. Gene expression of inducible nitric oxide synthase (iNOS), interferon (IFN)-γ, CXCL9 and IL-12p40 was increased in polyps of Apc Min/+ Ticam1 −/− mice. mRNA and protein expression of c-Myc, a critical transcription factor for inflammation-associated polyposis, were increased in polyps of Apc Min/+ Ticam1 −/− mice. A Lactobacillus strain producing dsRNA was detected in feces of Apc Min/+ mice.
Conclusion
These results imply that the TLR3/TICAM-1 pathway inhibits polyposis through suppression of c-Myc expression and supports long survival in Apc Min/+ mice.
Similar content being viewed by others
Background
Tumor progression is closely linked to inflammation [1]. The intestine contains microorganisms that influence the incidence of inflammation-associated cancer via toll-like receptors (TLRs) expressed in gut mucosal cells. TLR2/4 and Nod-like receptor (NLR)1 are expressed in mucosa and detect intestinal bacterial patterns [2]. Nod1 signal may ameliorate inflammation-induced polyposis through nuclear factor (NF)-κB and activator protein 1 (AP1). Nod1 is reportedly important for maintaining the integrity of the intestinal epithelium to protect it against injury, inflammation and subsequent carcinogenesis [3]. TLRs except for TLR3 activate the adaptor MyD88, which serves as a key factor for promotion of carcinogenesis and development in colon cancer [4, 5]. TLR2/4 respond to bacteria and modulate NF-κB activation during the inflammatory response in the TLR/MyD88 pathways [5]. Other receptors which activate MyD88 also participate in inflammation and tumorigenesis in intestinal epithelial cells [6, 7], suggesting a crucial role for MyD88 in homeostasis of intestine. Epithelial cells and microflora work together cooperatively to maintain mucosal homeostasis via MyD88 signaling.
Several reports have suggested that lactobacillus produces partial or structural double-stranded (ds) RNA, which can activate TLR3 in the intestine [8]. TLR3 is expressed in mucosal epithelial cells and myeloid cells, which may sample the bacterial by-products of dsRNAs via phagocytosis [9]. TLR3 couples with the adaptor TICAM-1 (TRIF) to activate transcription factors, IRF3 and AP1. If this is the case, both MyD88 and TICAM-1 pathways participate in polyposis under the presence of complex innate stimulation. To test the relationship between TLR3/TICAM-1 and intestinal polyposis, we employed the Apc Min/+ mouse model [10]. We found TICAM-1 is important for suppression of tumorigenesis and homeostasis of innate sensing of bacteria. We herein addressed the mechanism by which TLR3/TICAM-1 participates in polyp formation in Apc Min/+ mice.
Methods
Mice
Apc Min/+ mice on a C57BL/6 background were purchased from the Jackson Laboratory. Myd88 −/− C57BL/6 mice were provided from Dr. S Akira (Osaka University). Ticam1 −/− C57BL/6 mice were established in our laboratory. Apc Min/+ mice were crossed to Myd88 −/− or Ticam1 −/− mice to generate Apc Min+/− Myd88 +/− , Apc Min+/− Myd88 −/− , Apc Min+/− Ticam1 +/− and Apc Min/+ Ticam1 −/− littermates. Mice were bred and maintained under specific pathogen–free conditions. No abnormal behavior was observed in Apc Min/+ Ticam1 −/− during the period we maintained. In several individuals, growth retardation was observed for unknown reason, but we used individuals with normal body weight. Female and male mice were used for the present experiments. All animal experiments were approved by the University’s Committee on Use and Care of Animals.
Harvesting of polyps
Mice were sacrificed by cervical dislocation. The small and large intestines were harvested and washed with cold PBS. The intestines were longitudinally slit open to grossly count tumors with the aid of a magnifier and stereomicroscope. Polyps ≧2 mm were collected by forceps and used for the following experiments.
RT-qPCR and PCR
For quantitative PCR, total RNA was extracted with TRIzol, and 0.4 μg of RNA was treated with DNase I, and then reverse-transcribed using the High Capacity cDNA Transcription Kit (ABI) with random primers according to the manufacturer’s instructions. qPCR was performed using the Step One Real-Time PCR system (ABI). The RNA expression levels were normalized to Gapdh. To detect genomic DNA of Lactobacillus Johnsonii, DNA was extracted with QIAamp Stool Mini kit (Qiagen) form feces according to the manufacturer’s instructions. Purified genomic DNA was subjected to PCR using specific primers for Lactobacillus Johnsonii. PCR product was confirmed as amplified Lactobacillus Johnsonii genome by sequencing. The primers for detection of this bacillus were described in an early report [11]. Primer sequences used in this study are listed in Additional file 1: Table S1.
Immunofluorescence
The small intestines were fixed with 4% paraformaldehyde (PFA)/PBS for 1 h at 4 °C. Fixed tissues were impregnated with 15% sucrose/PBS for 1 h following 30% sucrose/PBS for overnight at 4 °C with rotation. Tissues were then embedded in O.T.C. compound (Sakura Finetek Japan) and the frozen tissue blocks were sectioned by using cryotome (LEICA CM1850). Sections were fixed with acetone on ice for 30 min. After three washes with PBS, the sections were blocked with mouse serum IgG in 5% BSA/PBS for 1 h at R.T. Sections were stained with FITC-labeled anti-CD11c, anti-CD11b, anti-CD4 or anti-CD8, and mounted with Prolong Gold (Thermo Fisher Scientific). Samples were monitored at × 63 or 40 magnification using an LSM510 META microscopy (Zeiss).
Flow cytometry
Single cell suspensions isolated from small intestine were stained with fluorescent dye-labeled Abs after blocking with an anti-CD16/32 Ab [12]. The following Abs were used: FITC- or APC/Cy7-CD45 (30-F11), PE-anti-CD11b (M1/70), APC-anti-CD11c (N418), APC-anti-CD3e (145-2C11), FITC-anti-CD4 (GK1.5), PE-anti-CD8a (53-6.7) (Biolegend). Dead cells were stained with 7AAD (Sigma). Samples were analyzed by a FACS Calibur or FACS Aria II (BD Bioscience); data analysis was performed using Flow Jo (Tree star).
SDS-PAGE/western blotting
Proteins were extracted from polyps by SDS-containing sample buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 35% glycerol, and BPB). The samples were resolved on SDS-PAGE (7.5 or 10% gel), and blotted onto PVDF membranes (Millipore). Proteins were detected by rabbit antibodies against c-Myc and GAPDH (Cell signaling technology). The blot was labeled with Horseradish peroxidase-conjugated goat Ab against rabbit Ig’s (Biosource). The color was developed by ECL Prime Western Blotting Detection Reagent (GE Healthcare).
Statistical analysis
P-values were calculated with Student t-test. Error bar represent the standard deviation (SD) between samples.
Results
Ticam1 deficiency results in short survival of Apc Min+ mice
TLR transmits signal through two major adaptors, MyD88 and TICAM-1 [13]. We generated Apc Min/+ mice on MyD88- or Ticam1-deficient background by crossing Apc Min/+ mice to Myd88 −/− or Ticam1 −/− mice, respectively. The average survival of Apc Min/+ mice was 28.6 weeks. Apc Min/+ Myd88 −/− mice survived longer than Apc Min/+ mice, consistent with previous reports [7]. In contrast, survival times for Apc Min/+ Ticam1 −/− mice were significantly shorter at ~20 weeks (Fig. 1a, b). Apc Min/+ Myd88 +/− and Apc Min/+ Ticam +/− mice showed similar life-spans, the average survival was 29 weeks indistinguishable from that of Apc Min/+ mice (Additional file 2: Figure S1), suggesting the Ticam1 gene disruption was the event that affected life span. Only homologous deficiency of MyD88 or TICAM-1 affected the survival time.
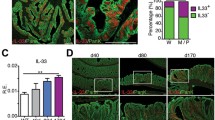
Multiple polyposis occurs in the intestine of Apc Min/+ mice, which represents intestinal tumorigenesis by adenomatous polyposis coli (APC) under the regulation of TLR signal [7]. Apc Min/+ Myd88 −/− mice reportedly have fewer polyps in the intestine than Apc Min/+ mice [7]. We then examined the polyp formation in Apc Min/+ Ticam1 −/− mice at 23~29 weeks of age (Fig. 2). Apc Min/+ mice showed a high frequency of polyp formation as reported previously [6, 7]. Apc Min/+ Ticam1 −/− mice had more polyps than Apc Min/+ mice in the distal small intestine (Fig. 2). Thus, the incidence of tumor formation is high in the middle and distal intestine of Apc Min/+ Ticam1 −/− mice compared to Apc Min/+ mice. Only a few polyps were observed in the proximal intestine and colon of Apc Min/+ mice, and no or minimal increases in polyp numbers was observed in Apc Min/+ Ticam1 −/− mice (Fig. 2).
The number of polyps is significantly increased in Apc Min/+ Ticam1 −/− mice. The small and large intestines were collected from 23 to 29-weeks-old mice. The small intestine was divided to three equal parts, proximal small intestine, middle and distal small intestine, and large intestine. Polyps (≧ 2 mm) were counted in the indicated mice. The graphs show the number of polyps in proximal small intestine, distal small intestine, and large intestine. Error bars show SD. p < 0.05 in Student’s t-test and n.s.; not significant
Immune cell infiltration in polyps
TLRs on epithelial cells recognize microbial products of commensal bacteria and induce inflammatory responses, including oncogene expression [14]. TLRs except TLR3 provoke MyD88 signaling and accelerate the proliferation of intestinal epithelial cells and prohibit apoptosis [15]. We focused on immune cells infiltrating into mucosal polyps. CD8 and CD4 mRNAs were minimally detected in unaffected mucosa in Apc Min/+ and were slightly increased in Apc Min/+ Ticam1 −/− mice at ~22 weeks age (Fig. 3a). CD11b and CD11c mRNAs levels were more increased in polyps of Apc Min/+ Ticam1 −/− mice than Apc Min/+ mice (Fig. 3a). mRNA for CD4 and CD8α, markers of myeloid cells, were also increased in Apc Min/+ Ticam1 −/− polyps (Fig. 3a). We detected more accumulation of CD8α-positive cells in polyps of Apc Min/+ Ticam1 −/− mice compared to Apc Min/+ mice by immunohistological staining (Fig. 3b). Similar tendencies were obtained with anti-CD11b antibody. The FACS profiles of the CD4-, CD8-, CD11b- and CD11c-positive cells in the intestine of Apc Min/+ Ticam1 −/− mice vs. Apc Min/+ mice are shown in Additional file 3: Figure S2. The mRNAs of these immune cells were only marginally increased in the normal (non-polyp) region of the small intestine in Apc Min/+ and Apc Min/+ Ticam1 −/− polyps (Additional file 4: Figure S3).
Immune cells infiltrate into polyps in Apc Min/+ Ticam1 −/− mice. a Gene expression in polyps prepared from 20 to 25-weeks-old mice were quantified by RT-qPCR. More than 3 mice in each group were used. **; p < 0.01 in Student’s t-test. b Immune staining of the small intestine prepared from 20 to 25-weeks-old mice using anti-CD11b and anti-CD8α antibodies. Data shows representative results of two independent experiments
Inflammatory parameters were also increased in the polyps of Apc Min/+ Ticam1 −/− mice (Fig. 4). mRNA expression of iNOS (Nos2) was highly induced in Apc Min/+ Ticam1 −/− polyps compared to Apc Min/+ polyps (Fig. 4). In addition, mRNA expression of CXCL9 (Cxcl9), IFN-γ (Ifng) and IL-12p40 (Il12p40) was slightly but significantly increased. In the normal (non-polyp) region, this tendency was not prominent (data not shown). Thus, tumor-related inflammation was induced to a greater extent in the intestine of Apc Min/+ Ticam1 −/− mice than in Apc Min/+ mice.
High expression of c-Myc in Apc Min/+ Ticam1 −/− polyps
Previous reports suggest that c-Myc mRNA is not increased in epithelial cells in response to MyD88 activation [7, 16]. Instead, β-catenin signaling is amplified by constitutive inactivation of APC and transcriptionally up-regulates the c-Myc mRNA [17]. We next checked the levels of the c-Myc mRNA in Apc Min/+ and Apc Min/+ Ticam1 −/− polyps. The levels of c-Myc mRNA were high in Apc Min/+ Ticam1 −/− polyps compared to Apc Min/+ polyps (Fig. 5a). The c-Myc protein was abundant in Apc Min/+ Ticam1 −/− polyps in comparison with Apc Min/+ polyps (Fig. 5b). The results were confirmed with confocal analysis (Fig. 5c). Although the staining density does not reflect the protein levels, the c-Myc protein level appears high in Apc Min/+ Ticam1 −/− polyps in Fig. 5c (and data not shown). The mRNA levels of PD-L1 (Pdl1) and COX2 (Ptgs2) appeared higher in c-Mychigh polyps than c-Myclow polyps. Conversely, expression of CD8α (Cd8a) and Perforin-1 (Prf1) in c-Mychigh polyps was lower than c-Myclow polyps. c-Mychigh polyps are likely to form a microenvironement favorable for tumor growth (Fig. 6). The TLR3 level was barely affected by environment in c-Myclow and c-Mychigh polyps (not shown): the genes affected by environment in c-Myclow vs. c-Mychigh polyps are shown in Additional file 5: Table S2.
High expression of c-Myc in polyps of Apc Min/+ Ticam1 −/− mice. a Gene expression in polyps prepared from 20 to 25-weeks-old mice were quantified by RT-qPCR. More than 3 mice in each group were used. **; p < 0.01 in Student’s t-test. b Protein was extracted from pooled three polyps in distal small intestine and separated by SDS-PAGE. Expression levels of c-Myc protein were detected by Western blotting. GAPDH is used as an internal control. Each lane shows results from pooled samples prepared from individual mice. Lane 1-2: polyps from Apc Min/+ mice, Lane 3-5: polyps from Apc Min/+ Ticam-1 −/− mice. c Immune staining of polyps in small intestine prepared from 20 to 25-weeks-old Apc Min/+ Ticam1 −/− mice using anti-c-Myc and anti-EpCAM antibodies. Data show representative results of two independent experiments
Differential gene expression in c-Myclow and c-Mychigh polyps. Polyps prepared from 20 to 25-weeks-old Apc Min/+ Ticam1 −/− mice were divided into two groups according to the high (≧ 2.0) and low (<2.0) expression levels of c-Myc and gene expression was compared by RT-qPCR. *; p < 0.05, **; p < 0.01 in Student’s t-test
Intestine of Apc Min/+ mouse contains bacteria with TLR3-stimulating capacity
Since TICAM-1 is the adaptor of TLR3 and TLR4 [5, 13], c-Myc may be suppressed by TLR3/4-TICAM-1 signaling. A previous report suggested that TLR3 is activated in response to dsRNA moieties yielded by Lactobacillus in mouse intestine [8]. PCR analysis using the specific primer sets detected the genome DNA of Lactobacillus johnsonii in the feces of Apc Min/+ mice, implying that the TLR3 signaling is constitutively activated in the intestine (Fig. 7).
Discussion
Carcinogenesis is established through multi-step gene mutations in intestinal epithelial cells. Loss-of-function of APC occurs in most patients of familial-associated polyposis [18] and causes malignant polyposis. Apc Min/+ mice have a mutation in the APC gene and accelerate polyposis in the intestine but not colon [10]. While Apc Min/+ mice die ~24 weeks from complication of tumorigenesis, their survival is prolonged by MyD88 disruption [7]. Thus, MyD88 signal of TLRs enhances protumor activity to shorten the survival. TICAM-1 transmits the other signal to activate a transcription factor IRF3. Here we showed that knockout of TICAM-1 results in short survival in Apc Min/+ mice, which suggests that the TLR3/TICAM-1 signal reduces polyposis promoted by the TLR2/4/MyD88 signaling pathway.
Intestinal epithelial cells express TLRs which utilize two adaptors, MyD88 and/or TICAM-1, as well as immune cells and tumor cells in mice [5, 13]. MyD88 evokes inflammatory signal that causes nuclear translocation of NF-κB and regulates apoptosis in tumor cells. Liberation of inflammatory cytokines sustains tumor-supporting microenvironment. In the Apc Min/+ mouse model of intestinal tumorigenesis, activation of the MyD88 pathway is related to stabilization of c-Myc protein but not to up-regulation of its mRNA in epithelial cells, resulting in a decrease in tumor growth in Apc Min/+ Myd88 −/− mice [7, 16]. On the other hand, the role of TICAM-1 in the regulation of c-Myc expression and tumorigenesis has been controversial [19, 20]. TICAM-1 has been identified in myeloid cells including dendritic cells and several subsets of macrophages [21,22,23]. The TLR3/TICAM-1 pathway takes part in cross-priming and IL-12 production that in turn causes DC priming and cytotoxic T cell (CTL) induction [22, 24, 25]. Moreover, some tumor cell lines express TLR3 [26]. We show that c-Myc expression is suppressed via TICAM-1: TICAM-1 signals constitutively suppress c-Myc expression and TICAM-1 loss results in c-Myc up-regulation. Over-expression of c-Myc abrogates its regulatory function in the cell cycle and induces tumorigenesis [17]. TLR3 signaling suppresses tumor cell growth through down-regulation of c-Myc [19]. The c-Myc level, however, barely affect the TLR3 expression. Thus, it would be reasonable to hypothesize that the TICAM-1 signaling pathway suppress c-Myc transcription and reduces intestinal polyp formation in Apc Min/+ mice. We have examined Ticam1-associated gene clusters by comprehensive method [27]. So, we selected inflammatory-induced genes form the Ticam1-associated genes (Additional file 5: Table S2). PolyI:C-activated TLR3-TICAM-1 signaling also suppresses tumor growth via immune activation [28]. Hence, TLR3 ligand may bimodally act on TLR3 expressed in tumor cells and immune cells, leading to tumor regression.
Our results imply that constitutive activation of the TLR3/TICAM-1 signaling pathway occurs in intestinal mucosa of Apc Min/+ mice. TLR3 stimulation also occurs with Lactobacillus dsRNA in the intestine [8]. In this scenario, bacterial-derived dsRNA behaves like a tumor suppressor via c-Myc regulation through the TLR3 signaling pathway. Thus, Lactobacillus may support good flora conditions to constitutively activate TLR3 in the intestinal epithelial cells or immune cells. TLR3/TICAM-1 signaling is likely to suppress c-Myc mRNA expression through direct stimulation of TLR3 on epithelial cells or by indirect stimulation via TLR3-expressing immune cells. Further study is required to elucidate the mechanism of TICAM-1-mediated suppression of c-Myc expression through intestinal microflora.
Many reports suggest that MyD88 induces protumor signal in tumor or transformed cells, but in dendritic cells MyD88 induces priming of T cells to regress tumor cells (28). Myeloid-derived suppressor cells and tumor-associated macrophages express TLR2 that activates MyD88 signaling to promote invasion and metastasis [29]. However, TLR3/TICAM-1 signals convert these myeloid cells to tumoricidal effectors in tumor microenvironment [30, 31]. Even in epithelial and tumor cells, stimulation of TLR3 does not promote cell growth or inflammation, which may be attributable to suppression of c-Myc. TLR3 adjuvant is now considered more successful in tumor immunotherapy compared to other TLR adjuvants. This study demonstrates an additional advantage of TLR3 adjuvant for direct therapeutic application to tumor cells: TLR3-targeted therapy may be of benefit to cancer patients by acting on both immune cells and tumor microenvironment.
Conclusion
The TLR3/TICAM-1 signaling suppresses c-Myc mRNA expression through direct stimulation of TLR3 in intestinal cells to suppress mucosal polyposis in Apc Min/+ mice. Survival time is shortened by knockout of Ticam-1 in Apc Min/+ mice.
Abbreviations
- AP1:
-
Activation protein 1
- APC:
-
Adenomatous polyposis coli
- CTL:
-
Cytotoxic T lymphocytes
- IFN:
-
Interferon
- IL:
-
Interleikin
- IRF:
-
Interferon regulatory factor
- MyD88:
-
Myeloid differentiation primary response gene 88
- Nod1:
-
Nucleotide-binding oligomerization domain-containing protein 1
- TICAM-1:
-
TIR domain-containing adapter molecule 1
- TLR:
-
Toll-like receptor
References
Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99.
Girardin SE, Tournebize R, Mavris M, et al. CARD4/Nod1 mediates NF-κB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001;2:736–42.
Chen YC, Shaw MH, Redondo G, Nunez G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 2008;68:10060–7.
Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41.
Seya T, Akazawa T, Uehori J, Matsumoto M, Azuma I, Toyoshima K. Role of toll-like receptors and their adaptors in adjuvant immunotherapy for cancer. Anticancer Res. 2003;23:4369–76.
Xiao H, Gulen MF, Qin J, Yao J, Bulek K, Kish D, Altuntas CZ, Wald D, Ma C, Zhou H, et al. The toll-Interleukin-1 receptor member SIGIRR regulates colonic epithelial homeostasis, inflammation, and tumorigenesis. Immunity. 2007;26:461–75.
Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–7.
Kawashima T, Kosaka A, Yan H, Guo Z, Uchiyama R, Fukui R, Kaneko D, Kumagai Y, You DJ, Carreras J, Uematsu S, Jang MH, Takeuchi O, Kaisho T, Akira S, Miyake K, Tsutsui H, Saito T, Nishimura I, Tsuji NM. Double-stranded RNA of intestinal commensal but not pathogenic bacteria triggers production of protective interferon-β. Immunity. 2013;38:1187–97.
Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–44.
Cooper HS, Everley L, Chang WC, et al. The role of mutant Apc in the development of dysplasia and cancer in the mouse model of dextran sulfate sodium-induced colitis. Gastroenterology. 2001;121:1407–16.
Zhang R, Daroczy K, Xiao B, Yu L, Chen R, Liao Q. Quantitative and semiquantitative analysis of lactobacillus species in the vaginas of healthy fertile and postmenopausal Chinese women. J Med Microbiol. 2012;61:729–39.
Shime H, Kojima A, Maruyama A, Saito Y, Oshiumi H, Matsumoto M, Seya T. Myeloid-derived suppressor cells confer tumor-suppressive functions on natural killer cells via polyinosinic:polycytidylic acid treatment in mouse tumor models. J Innate Immun. 2014;6:293–305.
Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76.
Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, Leiser A, Schwartz PE, Rutherford T, Mor G. Regulation of IKKβ by miR-199a affects NF-κB activity in ovarian cancer cells. Oncogene. 2008;27:4712–23.
Walsh MF, Ampasala DR, Hatfield J, Vander Heide R, Suer S, Rishi AK, Basson MD. Transforming growth factor-beta stimulates intestinal epithelial focal adhesion kinase synthesis via Smad- and p38-dependent mechanisms. Am J Pathol. 2008;173:385–99.
Gonzalez-Navajas J, Seo GS, Shen C, Herdman S, Varki N, Corr M, Lee J. ERK activation drives intestinal tumorigenesis in Apcmin/+ mice. Nat Med. 2010;16:665–70.
Wilkins JA, Sansom OJ. C-Myc is a critical mediator of the phenotypes of APC loss in the intestine. Cancer Res. 2008;68:4963–6.
Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70.
Pries R, Hogrefe L, Xie L, Frenzel H, Brocks C, Ditz C, Wollenberg B. Induction of c-Myc-dependent cell proliferation through toll-like receptor 3 in head and neck cancer. Int J Mol Med. 2008;21:209–15.
Lin LL, Huang CC, Wu CL, Wu MT, Hsu WM, Chuang JH. Downregulation of c-Myc is involved in TLR3-mediated tumor death of neuroblastoma xenografts. Lab Investig. 2016;96:719–30.
Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4:161–7.
Matsumoto M, Tatematsu M, Nishikawa F, Azuma M, Ishii N, Morii-Sakai A, Shime H, Seya T. Defined TLR3-specific adjuvant that induces NK and CTL activation without significant cytokine production in vivo. Nat Commun. 2015;6:6280.
Matsumoto M, Funami K, Tanabe M, Oshiumi H, Shingai M, Seto Y, Yamamoto A, Seya T. Subcellular localization of toll-like receptor 3 in human dendritic cells. J Immunol. 2003;171:3154–62.
Azuma M, Ebihara T, Oshiumi H, Matsumoto M, Seya T. Cross-priming for antitumor CTL induced by soluble Ag + polyI:C depends on the TICAM-1 pathway in mouse CD11c+ /CD8α+ dendritic cells. Oncoimmunology. 2012;1:581–92.
Azuma M, Takeda Y, Nakajima H, Sugiyama H, Ebihara T, Oshiumi H, Matsumoto M, Seya T. Biphasic function of TLR3 adjuvant on tumor and spleen dendritic cells promotes tumor T cell infiltration and regression in a vaccine therapy. Oncoimmunology. 2016;5:e1188244.
Matsumoto M, Seya T. TLR3: interferon induction by double-stranded RNA including poly(I:C). Adv Drug Deliv Rev. 2008;60:805–12.
Funami K, Matsumoto M, Oshiumi H, Obuse C, Seya T. The dataset of proteins specifically interacted with activated TICAM-1. Data Brief. 2016;8:697–9.
Seya T, Shime H, Takeda Y, Tatematsu M, Takashima K, Matsumoto M. Adjuvant for vaccine immunotherapy of cancer - focusing on TLR2 and TLR3 agonists for safely enhancing antitumor immunity. Cancer Sci. 2015;106:1659–68.
Kim S, Takahashi H, Lin WW, Descargues P, Grivennikov S, Kim Y, Luo JL, Karin M. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature. 2009;457:102–6.
Shime H, Matsumoto M, Oshiumi H, Tanaka S, Nakane A, Iwakura Y, Tahara H, Inoue N, Seya T. Toll-like receptor 3 signaling converts tumor-supporting myeloid cells to tumoricidal effectors. Proc Natl Acad Sci U S A. 2012;109:2066–71.
Shime H, Matsumoto M, Seya T. Double-stranded RNA promotes CTL-independent tumor cytolysis mediated by CD11b+Ly6G+ intratumor myeloid cells through the TICAM-1 signaling pathway. Cell Death Differ. 2017;24:385–96.
Acknowledgements
We are grateful to Drs. Kiyoshi Takeda and Eiji Umemoto (Osaka University, Osaka) for their kind support of this study. We thank Ms. N. Ishii-Mugikura, and A. Morii-Sakai for their technical support.
Funding
This work was supported in part by the Grants-in-Aid from the Ministry of Education, Science, and Culture (MEXT), “the Carcinogenic Spiral” a MEXT Grant-in-Project (T. Seya), the Ministry of Health, Labor, and Welfare of Japan (T. Seya, M. Matsumoto), Takeda Science Foundation (H. Shime), the Uehara Memorial Foundation, Smoking Research Foundation, and the Iskura Research Foundation (T. Seya).
Availability of data and materials
Not applicable.
Author information
Authors and Affiliations
Contributions
Design of the research, HS, MM, TS; Performing experiments, JO, HT, KT, KF, YT, SY; Data interpretation, HS, MM, TS; Writing manuscript, HS, MK, TS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
All animal research protocols for this work were reviewed and approved by the Animal Safety Center, Hokkaido University, Japan.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: Table S1.
Primer sequences used for real-time RT-PCR. (DOCX 19 kb)
Additional file 2: Figure S1.
Survival curve and days of ApcMin/+ mice. Survival curves (upper panel) and survival days (lower panel) were monitored in ApcMin/+, ApcMin/+Myd88−/+ and ApcMin/+Ticam1−/+ mice. N > 18 in each group. (PDF 2353 kb)
Additional file 3: Figure S2.
FACS analysis of immune cells in the small intestine. We checked the degrees of infiltration of immune cells into small intestine in ApcMin/+Ticam1−/− mice by FACS analysis. The whole small intestine was harvested from WT, ApcMin/+ and ApcMin/+ Ticam1−/− mice. The proportions of small intestine-infiltrating CD11b+, CD11c+, CD4+ T and CD8+ T cells were evaluated on FlowJo ver.9.9.4 (Tree Star). (PDF 456 kb)
Additional file 4: Figure S3.
Immune cell markers in the non-polyp region of the distal intestine. Gene expression in the non-polyp region of the distal intestine prepared from 20 to 25-weeks-old ApcMin/+ mice (n = 3) or ApcMin/+Ticam1−/− mice (n = 5) was quantified by RT-qPCR. n.s.; not significant in Student’s t-test. (PDF 68 kb)
Additional file 5: Table S2.
Relative expression levels of inflammatory-associated genes in c-Myclow and c-Mychigh polyps. (DOCX 19 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ono, J., Shime, H., Takaki, H. et al. The TLR3/TICAM-1 signal constitutively controls spontaneous polyposis through suppression of c-Myc in Apc Min/+ mice. J Biomed Sci 24, 79 (2017). https://doi.org/10.1186/s12929-017-0387-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12929-017-0387-z