Abstract
Background
Balanced structural variants are mostly described in disease with gene disruption or subtle rearrangement at breakpoints.
Case presentation
Here we report a patient with mild intellectual deficiency who carries a de novo balanced translocation t(3;5). Breakpoints were fully explored by microarray, Array Painting and Sanger sequencing. No gene disruption was found but the chromosome 5 breakpoint was localized 228-kb upstream of the MEF2C gene. The predicted Topologically Associated Domains analysis shows that it contains only the MEF2C gene and a long non-coding RNA LINC01226. RNA studies looking for MEF2C gene expression revealed an overexpression of MEF2C in the lymphoblastoid cell line of the patient.
Conclusions
Pathogenicity of MEF2C overexpression is still unclear as only four patients with mild intellectual deficiency carrying 5q14.3 microduplications containing MEF2C are described in the literature. The microduplications in these individuals also contain other genes expressed in the brain. The patient presented the same phenotype as 5q14.3 microduplication patients. We report the first case of a balanced translocation leading to an overexpression of MEF2C similar to a functional duplication.
Similar content being viewed by others
Background
Intellectual disability (ID) is a common disorder affecting up to 3% of the population [1]. Between 3 and 15% of patients with ID present numerical or structural chromosomal abnormalities mainly unbalanced rearrangements [2]. Only 0.6% of subjects carry an apparently balanced chromosomal rearrangement such as de novo reciprocal translocations [3].
The link between balanced rearrangements and ID can be explained by several mechanisms such as subtle rearrangement at the breakpoints [2, 4], perturbation of parental imprinting [5], disruption of one or two genes at the breakpoints leading to a loss of function of these genes [6], formation of a fusion gene with a novel function [7] or perturbation of gene expression (previously called positional effect) [8] and, more recently, changes in enhancers or DNA folding modifications within Topologically Associated Domains (TAD) [9, 10].
Separated by specific and robust boundaries, TADs restrict gene expression regulation inside them. Changes in enhancer - promoter interactions and breaking TAD boundaries have been reported to be pathogenic and “TADopathies” constitute an upcoming new category of human mendelian disease [11]. Recent studies showed that disruption in chromatin organization such as TADs can impact gene expression located distantly from breakpoint [12].
In this study, we report the molecular characterization of a de novo balanced reciprocal translocation t(3;5)(p26.3;q14.3) dn in a woman with ID. The breakpoint does not lead to the disruption of a gene but is localised 228-kb upstream of MEF2C gene on chromosome 5.
Case presentation
The proband is the first child of a healthy non-consanguineous couple. Medical family history showed a paternal niece with speech delay, a paternal half-sister with mild ID and a deceased paternal cousin with unspecified malformations.
The girl was born by vaginal delivery after an uneventful pregnancy. Birth parameters were at mean (birth weight: 3.200 kg; birth length: 49 cm; and occipital frontal circumference (OFC) 34 cm). She had global developmental delay diagnosed since 2 years old. She sat at 10 months and learned to walk at 22 months.
At 9 years old, she was diagnosed with attention deficit/hyperactivity disorder and delayed speech. Psychometric evaluation estimated her developmental stage at 3 years for a chronological age of 9 years. She has no autistic or stereotypic features and a unique febrile seizure episode.
Facial features include spread eyebrows, protruding ears with simplified helices and abnormal dermatoglyphics. She also had bilateral fifth finger clinodactyly as her father. Spectroscopic brain MRI, EEG, audition and visual explorations, abdominal ultrasound as well as skeletal X-rays were normal. Urine and blood metabolic screening were also normal.
The chromosome analysis of the patient and her parents reported a de novo apparently balanced reciprocal translocation 46,XX,t(3;5)(p26.3;q14.3)dn. FISH analysis with chromosome 3 and 5 painting probes showed the unique involvement of chromosomes 3 and 5 in this rearrangement (Fig. 1a).
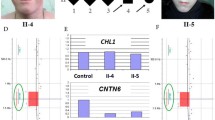
a GTG-banding chromosomes 3 and 5 and FISH nucleus assay. Black arrows show chromosome breakpoints on 3p26.3 and 5q14.3. A1. DAPI counterstain (blue). A2. Whole chromosome 3 painting probe (red). A3. Whole chromosome 5 painting probe (green). A4. Merging of A1, A2 and A3. b Predicted Hi C maps of der(5) from GM12878 cell line experiment (Liebermann -raw 10 kb) resolution. Black dashed line, yellow and grey bars represent predicted TAD. Blue genes & arrow are in chromosome 5 and green genes & arrow are in chromosome 3. CTCF sites are from ENCODE [13] data. c Expression of MEF2C gene in the patient’s lymphoblastoid cell lines (blue box) and three normal controls (green boxes), all assay were 3-times repeated, Y-axis shows the MEF2C RNA quantification normalized with the β-2 microglobulin housekeeping gene, ***: p < 0.001, One-way ANOVA with post-hoc Tukey HSD Test)
We confirmed the balanced status of the translocation using a microarray analysis which was normal (100 kb-resolution). Array painting assays and long-range PCR strategy allowed us to perform a fine mapping of these breakpoints. Breakpoints are located at chr3:920,589 and chr5:88,347,198 with the presence of a micro-homology of 3 nucleotides (TGC). No gene was interrupted in these regions. The chromosome 5 breakpoint is localized 228-kb upstream from ATG of the initiator codon of the MEF2C gene (NM_001193347). Visualisation of the 3D conformation using the 3D Genome Browser in 7 different cell types allow us to identify reliable TAD boundaries suggesting that the MEF2C gene and the LINC01226 long non coding RNA (lncRNA) exist in the same TAD on chromosome 5 [14]. The TAD on chromosome 3 contains only CNTN6 and CETN3 genes (Fig. 1b, Additional file 1: Figure S1 and Additional file 2: Figure S2). RNA studies revealed an overexpression of MEF2C in the patient’s lymphoblastoid cell line compared to 3 controls (gender- and age- matched with the patient) in experiments repeated three times (Fig. 1c). All genomic locations are based on Human Genome Build 37 (hg19).
Discussion and conclusions
Fine mapping of breakpoints on chromosomes 3 and 5 revealed no gene interruption but a breakpoint on the chromosome 5 localized 228-kb upstream of MEF2C.
The MEF2C gene causes the syndrome “Mental Retardation, Autosomal Dominant 20” (MIM # 613443) by haploinsufficiency [15]. Balanced translocation in this region have already been described in the literature. Such structural rearrangements on chromosome 5 create a single TAD encompassing MEF2C, resulting in decreased MEF2C expression [16]. The pathogenicity of MEF2C haploinsufficiency is no longer questioned to explain the phenotype of individuals with severe ID, stereotypic movement and autistic features. However, the pathogenicity of MEF2C overexpression is not clearly documented in the literature. Indeed, only 3 children and monochorionic diamniotic twins have been reported with a de novo 5q14.3 microduplication including MEF2C [17, 18] and MEF2C overexpression [19]. Interestingly, they share some pathological features such as global development delay with locomotor impairment (Table 1). Other genes included within these microduplications are also expressed in the brain. The major clinical sign described is a mild ID. Pathogenicity of MEF2C overexpression could be partly explained by its interaction on others genes known in human disease. Indeed, MEF2C overexpression could lead to MECP2 and CDKL5 upregulation [20]. MECP2 duplication in females is involved in psychiatric symptoms [21] and CDKL5 duplications are reported in women with heterogeneous symptoms, from learning difficulties to autistic behaviour, developmental delay, language impairment and hyperactivity [22].
In this article, we report the study of a patient who has ID associated with speech delay. According to the breakpoints of the translocation t(3;5), the predicted TAD in chromosome 3 only contains CNTN6 and CETN3 gene. Few studies described patients with ID carrying microdeletions/microduplications containing CNTN6 [23]. Still these CNVs have also been reported in some phenotypically normal individuals in the databases of genomic variants. They are mostly inherited from healthy parents and no patient has been identified with a point mutation of CNTN6 (ClinGen Dosage Sensitivity Map Curation https://www.ncbi.nlm.nih.gov/projects/dbvar/clingen/). To date CETN3 is not described in human disease. In chromosome 5, we identify a possible new TAD encompassing MEF2C and LINC01266. Our results of RNA quantification showed a clear significant overexpression of MEF2C in the patient’s lymphoblastoid cell line. Further FISH studies could be performed to completely confirm that MEF2C and LINC01266 are in the same TAD. LncRNAs are known to be involved in cis transcriptional regulation and chromosomal architecture [24]. According to GTEx, LINC01266 is also expressed in brain tissue [25]. No other major regulatory elements such as enhancers are predicted to be in this new TAD [26] . Localisation of the breakpoint is close to those of published cases, thus could not explain the upregulation (Additional file 1: Figure S1). As previously reported cases with balanced translocation around MEF2C all lead to a downregulation of the gene [16], our hypothesis is that LINC01266 could be involved in the upregulation of MEF2C.
To summarize, we report a disruption of chromatin organisation caused by balanced translocation t(3;5) with chromosome 5 breakpoint upstream of the overexpressed MEF2C gene, probably responsible for the patient’s phenotype. This case report adds substantial evidence of a specific phenotype associated with the overexpression of MEF2C.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. The main method descriptions is available in the Additional file 3.
Abbreviations
- der:
-
derivative chromosome
- EEG:
-
ElectroEncephaloGraphy
- ID:
-
Intellectual Disability
- kb:
-
kilobase
- Lnc:
-
Long non coding
- MRI:
-
Magnetic Resonance Imaging
- OFC:
-
Occipital Frontal Circumference
- t:
-
Translocation
- TAD:
-
Topologically Associated Domains
References
Vissers LELM, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18.
Schluth-Bolard C, Delobel B, Sanlaville D, Boute O, Cuisset J-M, Sukno S, et al. Cryptic genomic imbalances in de novo and inherited apparently balanced chromosomal rearrangements: array CGH study of 47 unrelated cases. Eur J Med Genet. 2009;52:291–6.
Rauch A, Hoyer J, Guth S, Zweier C, Kraus C, Becker C, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A. 2006;140A:2063–74.
Kumar A, Becker LA, Depinet TW, Haren JM, Kurtz CL, Robin NH, et al. Molecular characterization and delineation of subtle deletions in de novo “balanced” chromosomal rearrangements. Hum Genet. 1998;103:173 https://doi.org/10.1007/PL00008706.
Dupont J-M, Cuisset L, Cartigny M, Le Tessier D, Vasseur C, Rabineau D, et al. Familial reciprocal translocation t(7;16) associated with maternal uniparental disomy 7 in a Silver-Russell patient. Am J Med Genet. 2002;111:405–8.
Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006;79:370–7.
Di Gregorio E, Bianchi FT, Schiavi A, Chiotto AMA, Rolando M, di Cantogno LV, et al. A de novo X;8 translocation creates aPTK2-THOC2 gene fusion with THOC2 expression knockdown in a patient with psychomotor retardation and congenital cerebellar hypoplasia. J Med Genet. 2013;50:543–51.
Kleinjan DJ, van Heyningen V. Position effect in human genetic disease. Hum Mol Genet. 1998;7:1611–8.
Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–25.
Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538:265–9.
Lupiáñez DG, Spielmann M, Mundlos S. Breaking TADs: How Alterations of Chromatin Domains Result in Disease. Trends Genet. 2016;32:225–37.
Spielmann M, Lupiáñez DG, Mundlos S. Structural variation in the 3D genome. Nat Rev Genet. 2018. https://doi.org/10.1038/s41576-018-0007-0.
Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–801.
Wang Y, Song F, Zhang B, Zhang L, Xu J, Kuang D, et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19:151.
Vrečar I, Innes J, Jones EA, Kingston H, Reardon W, Kerr B, et al. Further Clinical Delineation of the MEF2C Haploinsufficiency Syndrome: Report on New Cases and Literature Review of Severe Neurodevelopmental Disorders Presenting with Seizures, Absent Speech, and Involuntary Movements. J Pediatr Genet. 2017;6:129–41.
Redin C, Brand H, Collins RL, Kammin T, Mitchell E, Hodge JC, et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet. 2017;49:36–45.
Novara F, Rizzo A, Bedini G, Girgenti V, Esposito S, Pantaleoni C, et al. MEF2C deletions and mutations versus duplications: a clinical comparison. Eur J Med Genet. 2013;56:260–5.
Le Meur N, Holder-Espinasse M, Jaillard S, Goldenberg A, Joriot S, Amati-Bonneau P, Guichet A, Barth M, Charollais A, Journel H, Auvin S, Boucher C, Kerckaert JP, David V, Manouvrier-Hanu S, Saugier-Veber P, Frebourg T, Dubourg C, Andrieux J, Bonneau D. MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J Med Genet. 2010;47(1):22–9.
Cesaretti C, Spaccini L, Righini A, Parazzini C, Conte G, Crosti F, et al. Prenatal detection of 5q14.3 duplication including MEF2C and brain phenotype. Am J Med Genet A. 2016;170A:1352–7.
Zweier M, Gregor A, Zweier C, Engels H, Sticht H, Wohlleber E, et al. Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum Mutat. 2010;31:722–33.
Ramocki MB, Peters SU, Tavyev YJ, Zhang F, Carvalho CMB, Schaaf CP, et al. Autism and other neuropsychiatric symptoms are prevalent in individuals with MECP2 duplication syndrome. Ann Neurol. 2009;66:771–82.
Szafranski P, Golla S, Jin W, Fang P, Hixson P, Matalon R, et al. Neurodevelopmental and neurobehavioral characteristics in males and females with CDKL5 duplications. Eur J Hum Genet. 2015;23:915–21.
Hu J, Liao J, Sathanoori M, Kochmar S, Sebastian J, Yatsenko SA, et al. CNTN6 copy number variations in 14 patients: a possible candidate gene for neurodevelopmental and neuropsychiatric disorders. J Neurodev Disord. 2015;7:26.
Tan JY, Smith AAT, Ferreira da Silva M, Matthey-Doret C, Rueedi R, Sönmez R, et al. cis-Acting Complex-Trait-Associated lincRNA Expression Correlates with Modulation of Chromosomal Architecture. Cell Rep. 2017;18:2280–8.
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5.
Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database. 2017;2017. https://doi.org/10.1093/database/bax028.
Acknowledgements
We deeply thank the patient and her family for their participation in this study. We thank Mrs. D. Mechin and Mrs. A. Fabre for sanger sequencing. We thank the microarray Core Facility of the Institute of Research on biotherapy, CHRU-INSERM-UM1 Montpellier, http://irb.chu-montpellier.fr and the CHROMOSTEM platform (http://www.chu-montpellier.fr/fr/chercheurs/plateformes/les-plateformes-recherche/chromostem/).
Funding
This work was funded by grants “Projet Hospitalier de Recherche Clinique” n° 7890 and “Appel d’Offre Interne” n°8688 from Regional University Hospital of Montpellier. These funds only supported the cost of reagents for all genetics tests except for array painting. They have no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
YK, SA and SS performed all patient analysis (except for array painting) and were major contributors in writing the manuscript. BLN performed the array painting analysis. GJB, CC, TM, GT, BP, WC, GD and PJ were involved in the genetic diagnosis and care. PF and GV coordinated the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not Applicable
Consent for publication
Parents gave their written consent to participate in this study and to publication, which was approved by the institutional ethics committee of Montpellier university hospital.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Figure S1. Localisation of breakpoints on Hi C maps from GM12878 cell line experiment on chromosome 3 and chromosome 5 (Liebermann -raw 10 kb resolution). Grey arrow represents the breakpoint localisation. Dashed blue arrow represent other breakpoint described by Redin et al. (Redin et al. [16]) with MEF2C downregulation. Blue genes & arrow are in chromosome 5 and green genes & arrow are in chromosome 3. (PNG 355 kb)
Additional file 2:
Figure S2. Chromosome 5 TAD boundaries across 4 different cell types (IMR90, NHEK, GM12878 and KBM7). Black dashed line, yellow and grey bars represent TADs. Grey arrow represents the breakpoint localisation. Black line represents TAD boundary. (PNG 257 kb)
Additional file 3
Materials and methods. (DOCX 15 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yauy, K., Schneider, A., Ng, B.L. et al. Disruption of chromatin organisation causes MEF2C gene overexpression in intellectual disability: a case report. BMC Med Genomics 12, 116 (2019). https://doi.org/10.1186/s12920-019-0558-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-019-0558-8