Abstract
Background
Bovine viral diarrhea virus (BVDV) is prevalent in Korean indigenous cattle, leading to substantial economic losses. This study was conducted to investigate the occurrence of BVDV. In 2016, a total of 143 blood samples were collected from asymptomatic Korean indigenous calves younger than 3-months of age from six different farms in the Republic of Korea (ROK).
Results
Eighty-seven calves (60.8%, 87/143) were tested positive for BVDV as evaluated by RT-PCR analysis. Phylogenetic analysis based on the 5′-untranslated region was used to classify these cases into three subtypes: BVDV-1b, BVDV-1o, and BVDV-2a. These results showed that BVDV-1b was the predominant subtype, while 2 samples clustered with BVDV-2a. Interestingly, one sample formed a separate group as a potentially new subtype, BVDV-1o. To our knowledge, this is the first report of BVDV-1o infection in Korean native calves. The BVDV-1o subtype identified in this study was closely related to cattle isolates obtained from Japan, indicating that this subtype is a new introduction to the ROK.
Conclusions
This study provides useful information for carrying out epidemiological surveys of BVDV in the ROK and developing a vaccine for future use in the ROK, particularly for the first detection of BVDV-1o in Korean indigenous calves. Further studies are required to investigate the prevalence and pathogenicity of this BVDV-1o subtype.
Similar content being viewed by others
Background
Bovine viral diarrhea virus (BVDV) causes significant economic losses worldwide in the cattle industry through decreased productive performance and immunosuppression of herds [1, 2]. BVDV belongs to the genus Pestivirus along with classical swine fever virus and border disease virus in the family Flaviviridae. BVDV infects not only cattle, but also pigs, goats, sheep, and wild ruminants. BVDV includes two species, BVDV-1 and BVDV-2. Additionally, HoBi-like pestivirus has been proposed as a third new species, BVDV-3. Based on the 5′-untranslated region (UTR), BVDV-1 can be further divided into 21 subtypes (1a−1u) [3], BVDV-2 into four subtypes (2a−2d) [4, 5], and BVDV-3 into two genotypes of Brazilian and Thai origin [6]. Infection with BVDV is characterized by gastroenteritis, respiratory diseases and reproductive problems including abortion, congenital abnormalities, and the development of persistently infected (PI) calves that gain immunotolerance to BVDV through vertical transmission of the virus during early gestation [1, 7, 8].
To date, seven BVDV subtypes (1a, 1b, 1c, 1d, 1 m, 1n, and 2a) have been identified in the Republic of Korea (ROK) [9, 10]. Of these, BVDV-1b and BVDV-2a are predominant and widespread in cattle in the ROK [10,11,12]. The genetic diversity of BVDV must be considered when designing and constructing effective vaccination strategies against the virus. Additionally, a vaccine must accurately reflect the antigenic subtypes present in the country of use [13]. In the present study, we report a new subtype recently identified in the ROK. The results provide useful information for vaccine development.
Methods
Sample collection
Blood samples were collected from 143 asymptomatic Korean indigenous calves under 3 months of age from six different farms (Gimje, Gochang, Iksan, Sancheong, Wanju, and Hoenseong) in the ROK (Fig. 1). Five milliliters of blood was collected from the jugular veins of each calf in EDTA-supplemented tubes. The samples were delivered to the lab immediately after blood collection. These calves were the offspring of cows that had not previously been vaccinated against BVDV. The collected samples were stored at −80°C until analysis.
RNA extraction, RT-PCR, and sequencing
Total RNA was extracted from 200 μL of blood using RNAiso Plus Reagent (Takara Bio, Shiga, Japan) according to the manufacturer’s instructions. RNA was eluted in 20 μL of RNase-free water and stored at −80°C. The 5′-UTR and N-terminal protease (Npro) regions were used to detect BVDV as previously described [4, 11]. Reverse transcription-polymerase chain reaction (RT-PCR) was performed to amplify BVDV using DiaStar™ One-Step RT-PCR Smart Mix (Solgent, Daejeon, Korea). Briefly, RT-PCR was carried out at 50°C for 30 min, followed by 94°C for 5 min, and then 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, with a final extension step at 72°C for 10 min. Distilled water was used as a negative control in each PCR run. The predicted sizes of the amplified PCR products were 288 bp and 425 bp for the 5′-UTR and for Npro, respectively. The amplicons were subjected to 1.5% agarose gel electrophoresis and visualized following ethidium bromide staining. The PCR products were purified using an AccuPrep® PCR Purification Kit (Bioneer, Daejeon, Korea) and cloned into the pGEM®-T Easy vector (Promega, Madison, WI, USA), which was directly sequenced (Macrogen, Inc., Seoul, Korea).
Phylogenetic analysis
For homology analysis of BVDV genes, the obtained sequence data were analyzed using the Basic Local Alignment Search Tool of the National Center for Biotechnology Information database. Homologous sequences were analyzed using Chromas software (version 2.33, http://www.technelysium.com.au/chromas.html) and aligned using ClustalX (version 1.8). Construction of the phylogenetic trees based on the 5′-UTR and Npro were performed using the maximum-likelihood method with a Kimura 2-parameter [14] substitution model using MEGA7 software [15]. To construct each phylogenetic tree, additional sequences were obtained from GenBank (http://www.ncbi.nlm.nih.gov). The 5′-UTR of the gene sequences obtained in this study was assigned the following accession numbers: MH355922−MH355950, MH396616, and MH396618 for BVDV-1b, and MH396617 and MH396619 for BVDV-2a. The 5′-UTR and Npro for the gene sequence of BVDV-1o obtained in this study were assigned the accession numbers MF449421 and MG030482, respectively.
Statistical analysis
Statistical analysis was performed to assess the association between BVDV infection and the variables: colostrum intake and grazing. Binary univariate logistic regression models were constructed using the generalized linear mixed model in SPSS 24.0 software package (SPSS, Inc., Chicago, IL, USA), with the farm as a random effect. Odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to assess the likelihood of association. A value of P < 0.05 was considered significant.
Results
From the analysis of 143 blood samples, BVDV was detected in 87 calves (60.8%) from five of the six farms sampled (Table 1). Data for colostrum intake and growth type from the farms in the regions where fecal samples were collected are presented in Table 1. The Korean native calves examined in this study were born to cows that were not vaccinated against BVDV. There was no significant difference between colostrum intake and BVDV infection (P = 0.09). In calves that had not been fed colostrum (Heongseong and Iksan), BVDV infection was reduced by grazing (Iksan). In the Iksan and Sancheong regions where animals are grazed, colostrum-fed calves (Sancheong) showed an even higher rate of BVDV infection than in Iksan, where calves had not been fed colostrum (Table 1). Our results show that grazing decreased the OR for the incidence of BVDV by 5-fold compared to housing calves (OR = 0.2; 95% CI: 0.1−0.4; P = 0.001). BVDV infection showed the highest prevalence (64.7%) in 1−21-day-old animals and the prevalence of BVDV infection gradually reduced with age, but remained high (Table 2).
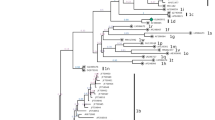
To investigate the genetic relationship between the BVDV isolates identified in this study, we sequenced all amplicons. Of these, 34 good sequences for 5′-UTR and one sequence for Npro were obtained and were included in the phylogenetic tree. Thirty-four isolates identified from Gimje, Gochang, Sancheong, Wanju, and Hoengseong were classified into 3 subtypes: BVDV-1b (31 isolates), BVDV-1o (1 isolate, new subtype), and BVDV-2a (2 isolates) based on sequence analysis of 5′-UTR (Fig. 2). Particularly, in case of Hoengseong farm, three subtypes were detected simultaneously. Phylogenetic analysis revealed genetic differences in samples derived from the same farm. Two isolates of BVDV-2a were detected in this study and were similar to those that had previously been isolated in the ROK. The BVDV-1o found in Hoengseong farm was closely related to that identified in Japanese cattle (LC089875) (Fig. 2). Another phylogenetic tree based on the Npro region was constructed to confirm the subtyping of BVDV-1o. As shown in Fig. 3, the Hoengseong isolate formed the same branch as BVDV-1o as determined by the sequence of 5′-UTR. Additionally, higher similarity was observed for most isolate relationships, except for the camel isolate. Homology values between the Hoengseong isolate identified in this study and BVDV-1o reference strains LC089875 and AB359931 were 95 and 88% for the 5′-UTR sequence and Npro sequence, respectively. This is the first report of BVDV-1o infection in calves in the ROK. The results demonstrated that BVDV-1b was the most frequently detected subtype and genetic variations were evident among BVDV-1b circulating in the ROK (Fig. 2).
Phylogenetic tree based on partial nucleotide sequences of the 5′-UTR of reference BVDV strains/isolates and Korean isolates identified in this study was constructed in MEGA7 using the maximum-likelihood (ML) method. The robustness of the tree was evaluated by bootstrapping 1000 replicates by ML. The Korean isolates identified in this study are indicated in bold
Discussion
In this study, we investigated the occurrence of BVDV in Korean indigenous calves. Our findings showed that the infection rate of BVDV was high, indicating the difficulty in controlling the spread of BVDV in the field. Unexpectedly, the highest rate of BVDV infection was observed in 1−21-day-old calves. Most BVDV infections in calves less than 30-days-old are likely PI. In this study, we could not examine whether these calves were PI or acutely infected because the farmers objected to a second collection of blood from the calves. Previous reports from several countries have highlighted the clinical significance of BVDV infections, particularly of PI [16, 17]. Because PI animals remain unidentified, they can contribute to the spread of BVDV to other members of the herd. Measures to prevent BVDV circulation in herds should focus on removing the PI animals. Alterations in the immune system due to BVDV infection result in an increased susceptibility to other enteric diseases in PI calves [18, 19]. Because PI animals are important for the ongoing transmission of BVDV in a population, our findings highlight the need for rapid diagnosis and elimination of PI animals.
Our findings showed that BVDV infection was not associated with colostrum intake (P = 0.09). However, BVDV infection significantly decreased when animals were allowed to graze in a pasture (P = 0.001) rather than being confined to housing. In this study, BVDV infection was not detected in the Iksan region, where animals grazed. Interestingly, on the Sancheong farm, where animals also grazed, the prevalence of BVDV was relatively higher than that on the Iksan farm (Table 1). This difference between the two farms may be related to colostrum intake. The prevalence of BVDV was high in colostrum-fed calves, supporting the possibility that cows that served as sources of colostrum were infected with BVDV or PI. Moreover, the high prevalence of BVDV in calves can be explained by the failure to vaccinate. In the ROK, vaccination of pregnant dams against BVDV is not implemented on most farms. This study highlights the importance of BVDV vaccination. Consequently, an outbreak of BVDV may commonly be associated not only with grazing but also with other factors, including cattle management systems, herd sizes, and vaccination. Further studies are necessary to determine the effects of these variables in reducing the occurrence of BVDV.
Although the number of samples was limited, our results revealed that three subtypes (BVDV-1b, BVDV-1o, and BVDV-2a) of BVDV were detected in Korean native cattle. Of these, BVDV-1b was the predominant subtype identified. Our data support those of previous studies [10, 11]. However, the BVDV isolates obtained in this study showed more genetic variation compared to those in other Korean BVDV-1b strains/isolates reported previously in the ROK. Additionally, two isolates were classified as BVDV-2a based on phylogenetic analysis. The BVDV-2a outbreak is known to be associated with severe acute infections in the ROK [12], but the isolates were detected in calves without any clinical symptoms. Recent studies demonstrated that BVDV-2c causes severe and often fatal illness in cattle in European countries [20, 21]. Thus, disease outbreaks appear to change over time. BVDV-1o was identified for the first time in the present study, and a total of eight subtypes of BVDV (1a, 1b, 1c, 1d, 1 m, 1n, 1o, and 2a) have now been found in the ROK, including BVDV-1o newly detected. This study provides useful information for constructing a vaccine for future use in the ROK, particularly for detecting the BVDV-1o subtype in Korean indigenous calves. BVDV1o was first isolated from a calf that developed a mucosal disease and from PI calves in Japan [22, 23], and has been detected in camels, goats, and pigs in China [3, 24, 25]. However, this isolate has not been detected anywhere other than Japan and China. Our results indicate that BVDV-1o is genetically heterogeneous but geographically restricted. At present, we cannot conclude how this virus spread. It is possible that trade between countries could be a possible pathway for the introduction of the new subtype into Korean cattle.
Conclusions
Our results provide evidence of the predominance of BVDV-1b in Korean native calves. Additionally, we identified the BVDV-1o subtype, which had not been previously documented in the ROK. As the number of calves examined was limited, larger epidemiological surveys are needed to investigate the prevalence of the BVDV-1o subtype as well as other subtypes. These results provide useful information for the development an effective vaccination for BVDV control.
Abbreviations
- 5′-UTR:
-
5′- untranslated region
- BVDV:
-
Bovine viral diarrhea virus
- ML:
-
Maximum-likelihood
- Npro :
-
N-terminal protease
- PI:
-
Persistent infection
- ROK:
-
Republic of Korea
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
References
Gates MC, Humphry RW, Gunn GJ, Woolhouse ME. Not all cows are epidemiologically equal: quantifying the risks of bovine viral diarrhoea virus (BVDV) transmission through cattle movements. Vet Res. 2014;45:110.
Pinior B, Firth CL, Richter V, Lebl K, Trauffler M, Dzieciol M, Hutter SE, Burgstaller J, Obritzhauser W, Winter P, et al. A systematic review of financial and economic assessments of bovine viral diarrhea virus (BVDV) prevention and mitigation activities worldwide. Prev Vet Med. 2017;137:77–92.
Mao L, Li W, Yang L, Wang J, Cheng S, Wei Y, Wang Q, Zhang W, Hao F, Ding Y, et al. Primary surveys on molecular epidemiology of bovine viral diarrhea virus 1 infecting goats in Jiangsu province, China. BMC Vet Res. 2016;12:181.
Vilcek S, Paton DJ, Durkovic B, Strojny L, Ibata G, Moussa A, Loitsch A, Rossmanith W, Vega S, Scicluna MT, et al. Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Arch Virol. 2001;146:99–115.
Flores EF, Ridpath JF, Weiblen R, Vogel FS, Gil LH. Phylogenetic analysis of Brazilian bovine viral diarrhea virus type 2 (BVDV-2) isolates: evidence for a subgenotype within BVDV-2. Virus Res. 2002;87:51–60.
Liu L, Xia H, Wahlberg N, Belak S, Baule C. Phylogeny, classification and evolutionary insights into pestiviruses. Virology. 2009;385:351–7.
Brownlie J, Clarke MC, Howard CJ. Experimental production of fatal mucosal disease in cattle. Vet Rec. 1984;114:535–6.
Polak MP, Zmudzinski JF. Experimental inoculation of calves with laboratory strains of bovine viral diarrhea virus. Comp Immunol Microbiol Infect Dis. 2000;23:141–51.
Joo SK, Lim SI, Jeoung HY, Song JY, Oem JK, Mun SH, An DJ. Genome sequence of bovine viral diarrhea virus strain 10JJ-SKR, belonging to genotype 1d. Genome Announc. 2013;1:4.
Oem JK, Chung JY, Roh IS, Kim HR, Bae YC, Lee KH, Jin YH, Lee OS. Characterization and phylogenetic analysis of bovine viral diarrhea virus in brain tissues from nonambulatory (downer) cattle in Korea. J Vet Diagn Invest. 2010;22:518–23.
Choi KS, Song MC. Epidemiological observations of bovine viral diarrhea virus in Korean indigenous calves. Virus Genes. 2011;42:64–70.
Oem JK, Hyun BH, Cha SH, Lee KK, Kim SH, Kim HR, Park CK, Joo YS. Phylogenetic analysis and characterization of Korean bovine viral diarrhea viruses. Vet Microbiol. 2009;139:356–60.
Mahony TJ, McCarthy FM, Gravel JL, Corney B, Young PL, Vilcek S. Genetic analysis of bovine viral diarrhoea viruses from Australia. Vet Microbiol. 2005;106:1–6.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Graham DA, Clegg TA, O'Sullivan P, More SJ. Influence of the retention of PI calves identified in 2012 during the voluntary phase of the Irish national bovine viral diarrhoea virus (BVDV) eradication programme on herd-level outcomes in 2013. Prev Vet Med. 2015;120:298–305.
Kelling CL, Steffen DJ, Cooper VL, Higuchi DS, Eskridge KM. Effect of infection with bovine viral diarrhea virus alone, bovine rotavirus alone, or concurrent infection with both on enteric disease in gnotobiotic neonatal calves. Am J Vet Res. 2002;63:1179–86.
Gunn GJ, Stott AW, Humphry RW. Modelling and costing BVD outbreaks in beef herds. Vet J. 2004;167:143–9.
Hessman BE, Fulton RW, Sjeklocha DB, Murphy TA, Ridpath JF, Payton ME. Evaluation of economic effects and the health and performance of the general cattle population after exposure to cattle persistently infected with bovine viral diarrhea virus in a starter feedlot. Am J Vet Res. 2009;70:73–85.
Gethmann J, Homeier T, Holsteg M, Schirrmeier H, Sasserath M, Hoffmann B, Beer M, Conraths FJ. BVD-2 outbreak leads to high losses in cattle farms in western Germany. Heliyon. 2015;1:e00019.
Jenckel M, Hoper D, Schirrmeier H, Reimann I, Goller KV, Hoffmann B, Beer M. Mixed triple: allied viruses in unique recent isolates of highly virulent type 2 bovine viral diarrhea virus detected by deep sequencing. J Virol. 2014;88:6983–92.
Nagai M, Hayashi M, Itou M, Fukutomi T, Akashi H, Kida H, Sakoda Y. Identification of new genetic subtypes of bovine viral diarrhea virus genotype 1 isolated in Japan. Virus Genes. 2008;36:135–9.
Sato A, Tateishi K, Shinohara M, Naoi Y, Shiokawa M, Aoki H, Ohmori K, Mizutani T, Shirai J, Nagai M. Complete genome sequencing of bovine viral diarrhea virus 1, subgenotypes 1n and 1o. Genome Announc. 2016;4:1.
Deng Y, Sun CQ, Cao SJ, Lin T, Yuan SS, Zhang HB, Zhai SL, Huang L, Shan TL, Zheng H, et al. High prevalence of bovine viral diarrhea virus 1 in Chinese swine herds. Vet Microbiol. 2012;159:490–3.
Gao S, Luo J, Du J, Lang Y, Cong G, Shao J, Lin T, Zhao F, Belak S, Liu L, et al. Serological and molecular evidence for natural infection of Bactrian camels with multiple subgenotypes of bovine viral diarrhea virus in western China. Vet Microbiol. 2013;163:172–6.
Funding
This work was supported by the National Research Foundation of Korea (NRF), funded by the Korea government (MSIP) (No. 2015R1C1A2A01053080).
Availability of data and materials
The datasets analyzed within the current study are available from the corresponding author upon request.
Author information
Authors and Affiliations
Contributions
KSC designed this study and wrote the manuscript. DGH and JHP participated in sample collection. DGH and JHR performed the experiments, data analysis, and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Animal handling and all experiments were approved by the animal ethics and welfare committee of Chonbuk National University (approval number 2016–00026). Written informed consent was provided by cattle owners for the collection of blood samples by an experienced and practicing veterinarian.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Han, DG., Ryu, JH., Park, J. et al. Identification of a new bovine viral diarrhea virus subtype in the Republic of Korea. BMC Vet Res 14, 233 (2018). https://doi.org/10.1186/s12917-018-1555-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-018-1555-4