Abstract
Background
Echinococcus granulosus sensu lato (s.l.) is the causative agent of cystic echinococcosis (CE), which is a cosmopolitan zoonotic parasitic disease infecting humans and a wide range of mammalian species including cattle. Currently, little information is available on the genetic diversity of Echinococcus species among livestock in Sudan. In the present study, fifty (n = 50) hydatid cysts were collected from cattle carcasses (one cyst sample per animal) at Al-kadarou slaughterhouse, Khartoum North, Sudan. DNA was extracted from protoscolices and the germinal layer of each cyst and subsequently amplified by PCR targeting the mitochondrial NADH dehydrogenase subunit 1 (NADH-1) gene. The amplified PCR products were purified and subjected to direct sequencing for subsequent construction of phylogenetic tree and net work analysis.
Results
The phylogenetic tree revealed the presence of Echinococcus canadenesis genotype 6 (G6) in 44 cysts (88.0%), Echinococcus ortleppi genotype 5 (G5) in 4 cysts (8.0%) and Echinococcus granulosus sensu stricto (s.s) genotype 1 (G1) in 2 cysts (4.0%). The phylogenetic network analysis revealed genetic variation among the different haplotypes/genotypes. This report has provided, for the first time, an insight of the role of cattle in the transmission of the zoonotic G1 echinococosis.
Conclusions
The results of the study illustrate that Sudanese breeds of cattle may play an important role in the transmission dynamics and the epidemiology of cystic echinococcosis in Sudan. This study reports the first molecular identification of E. granulosus s.s. in cattle in Central Sudan.
Similar content being viewed by others
Background
Cystic echinococcosis (CE) is a significant public health problem with high endemicity in east and central Africa including Sudan [1,2,3,4]. The larval stage of Echinococcus granulosus sensu lato (s.l.) causes CE in humans and a wide range of mammalian species. The life cycle involves the ingestion of parasite eggs by an intermediate host belonging to wildlife and domestic livestock species, including cattle. The dog is considered as the definitive host for this parasitic infection [5]. Humans are accidental dead end hosts. It is estimated that CE results in economic losses in the livestock sector due to morbidity. In addition, partial or total condemnations of infected organs of slaughtered animals are frequently encountered in endemic areas [5,6,7,8,9]. Echinococcosis has recently been included by the World Health Organization (WHO) as a neglected tropical disease [10]. CE may significantly affect the overall development and work productivity in endemic areas. In pastoral Sudanese communities, CE remains highly endemic with higher prevalence compared to agricultural communities. CE is endemic in most parts of the world, including regions of South America, the Mediterranean, Eastern Europe, East Africa, the Near and Middle East, Central Asia, China and Russia [7, 10,11,12,13]. Currently, ten distinct genotypes of E. granulosus s.l., designated as G1-G10, have been described worldwide on the basis of genetic diversity related to nucleotide sequences of the mitochondrial NADH dehydrogenase subunit 1 (NADH 1) and cytochrome C oxidase subunit 1 (COX1) genes. These different genotypes are associated with distinct intermediate hosts including sheep, goats, horses, cattles, pigs, camels and and members of the cervid family [14,15,16,17,18,19]. Of the ten genotypes of E. granulosus s.l., the cattle (G5) and the camel (G6) strains have already been reported among humans and livestock in Sudan [2, 20, 21]. Recent epidemiological studies indicated that the camel genotype (G6) was the most prevalent strain in Sudan [4, 22]. The extensive intra-specific genetic variation of E. granulosus s.l. could be better understood within the context of variations in the life cycle pattern [23, 24]. It is suggested that, different genotypes would probably exhibit different antigenicity, transmission profiles, pathological consequences, and different sensitivity to chemotherapeutic agents [25]. A lot of research efforts have been directed towards the epidemiology of CE in Sudan [26,27,28,29]. However, only few reports of the genetic diversity of the parasite among the cattle in Sudan employed sequence analysis of mitochondrial markers [2, 4, 21]. It is, therefore, becoming increasingly obvious that expanding the existing sequence data on the genetic diversity of E. granulosus s.l. is necessary to better understand the biology, ecology and molecular epidemiology of this parasite. In this investigation, a molecular characterization was conducted to identify hydatid cysts recovered from local cattle breed in Central Sudan.
Methods
Collection of samples and processing
Fifty hydatid cysts (n = 50) were collected over a period of 6 months from cattle during April–October, 2016, at Al-kadarou slaughterhouse, Khartoum North, Central Sudan. This slaughter house is the major cattle battoir in Khartoum North, Sudan. Hydatid cysts were obtained from cattle instantly after slaughtering and transferred in thermo-flasks to the Molecular Biology Laboratory at the Faculty of Veterinary Medicine, University of Khartoum, for processing and molecular characterization.
DNA extraction from hydatid cysts
Parasite genomic DNA was extracted from hydatid cysts as described by Ahmed and his coworkers [4]. Maximum DNA yield was obtained by spinning at 12,000 rpm for 1 min at room temperature. From the suspended nucleic acid, 5 μl was used in the PCR amplification.
Primers design and PCR assays
The primers were designed based on the published sequences of NADH dehydrogenase subunit 1 (NADH-1) gene of E. granulosus genotype 6 (G6) reported by Bowles and McManus [15]. Briefly, primer EGL1: 5′TGA AGT TAG TAA TTA AGT TTA A′3 and primer EGR2: 5′AAT CAA ATG GAG TAC GAT TA′3 were designed to amplify a fragment of 435 bp of E. granulosus s.l. by PCR. The details of PCR amplification, visualization and of results were described previously [4].
Sequence processing and phylogenetic analysis
The PCR products were purified using QIAquick PCR purification kit (Felden, Germany) and submitted for sequencing to a commercial company (Macrogen, Seoul, Korea). Bidirectional sequence fragments of the forward and reverse primers were generated for each sample. These were edited manually to correct possible base calling errors using BIOEDIT 7.0 and were subsequently joined to reconstruct a fragment of 344 bp of the parasite (NADH-1) gene. The consensus sequences were aligned with the corresponding region of NADH-1 gene of known genotypes circulating globally using CLUSTAL-X 2.1 [30]. The phylogenetic tree was constructed using the unweighted pair group method with arithmetic mean (UPGAM) implemented in MEGA software version 6.0 with 1000 bootstrap replicates [31]. Corresponding nucleotide sequences of NADH-1 of Taenia multiceps with GenBank accession number HM143887 were used as out groups in the constructed phylogenetic trees.
Phylogenetic network analysis
To measure the genetic variability, the number of haplotypes was determined using DNASP v5 [31] with insertions and deletions considered as variable sites. We used the median-joining (MJ) network algorithm [32] implemented in NETWORK 4.6 (www.fluxus-engineering.com).
Results and discussion
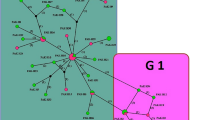
Microscopic examination revealed that all hydatid cysts were fertile and measured 2–10 cm in diameter. The predilection sites of the cysts were found to be the lung and the liver. All fifty DNA samples were amplified by PCR and generated a fragment of 435 bp of the NADH-1 gene. The partial sequences of the NADH-1 gene representing genotypes G1 (accession number LC167080), G5 (accession number LC167081) and G6 (accession numbers LC167082 and LC167083) were submitted to GenBank, DNA Data Base of Japan (DDBJ). The sequence analysis indicated a prevalence of (88.0%, n = 44), (8.0%, n = 4), (4.0%, n = 2) for Echinococcus canadenesis (G6), Echinococcus ortleppi (G5), and E.granulosus sensu stricto s.s (G1), respectively. The phylogenetic network analysis revealed clear genetic variation between the different genotypes and haplotypes. The present investigation indicated that at least three different genotypes of E. granulosus s.l. are actively circulating in cattle in Sudan as illustrated by the phylogenetic tree (Fig. 1) and phylogenetic network analyses (Fig. 2). The sample Sudan HC1_HP was well grouped with haplotype 6 and samples SudanHC2_HP1 that was clustered with genotype 6 in the phylogenetic tree was six SNPs different from the haplotype 6. Sudan 40_HP was grouped with genotype 1 and differed with only 2 SNPs from the previously known genotype 1.The three Echinococcus genotypes (G1, G5 and G6) reported in this study are all known human pathogens of significant public health concern [33]. The exclusive occurrence and a predominant circulation of the camel genotype (G6) in the bovine species suggested that cattle can play an important role in the transmission dynamic and the epidemiology of the disease [4]. The present study indicated that E. granulosus s.s., the sheep strain (G1), should equally be considered as an important infectious form of CE among cattle in Central Sudan.
Phylogenetic relationship of hydatid cysts of Echinococcus granulosus sensu lato recovered from Sudanese cattle and other genotypes identified globally. NADH dehydrogenase subunit 1 (NADH-1) partial sequences generated from this study were aligned with sequences of other strains from different parts of the world. Sequences were analyzed with the BioEdit software (Ibis Biosciences, Carlsbad, CA, USA). The phylogenetic tree was constructed using unweighted pair group method with arithmetic mean (UPGMA) implemented in MEGA software version 6.0 [31]. Bootstrap values were calculated from analysis of 500 replicates of the data set, and values greater than 50% are indicated at the appropriate nodes. Each genotype was designated by its GenBank accession number and the country of origin when available. The GenBank accession numbers (LC167080, LC167081) were given for Echinococcus granulossus sensu stricto (G1) and Echinococcus orteleppi (G5), respectively. Echinococcus canadensis genotypes (G6) were given accession numbers LC167082 and LC167083). Corresponding nucleotide sequence of NADH 1 of Taenia multiceps, GenBank accession number HM143887, was used as an out group. The partial NADH-1gene sequences identified in this study were highlighted in red color for clarity of the constructed phylogenetic tree
Phylogenetic network analysis of haplotypes. The number of haplotypes was determined with insertions and deletions considered as variable sites. Median-joining (MJ) network algorithm [32] implemented in NETWORK 4.6 was used to construct the phylogenetic network. The GenBank accession numbers were the same as indicated for the phylogenetic tree
Conclusions
The present study represents the first molecular record of E. granulosus s.s G1, thus reinforcing its role as a source of infection among Sudanese cattle breeds. In addition, this investigation provides additional information on the existing data indicating that Echinococcus granulosus s.s. G1, which was previously restricted to other region in the African continent, is now becoming broadly distributed in the country. Active surveillance is required to determine the distribution and prevalence of CE and to identify the genotypes/strains circulating in different regions of Sudan.
Abbreviations
- CE:
-
Cystic echinococcosis
- cox1:
-
Cytochrome C oxidase subunit 1
- DDBJ:
-
DNA Data Base of Japan
- E.granulosus s.l. :
-
Echinococcus granulosus sensu lato
- E.granulosus s.s. :
-
Echinococcus granulosus sensu strict: s.s
- G1:
-
Genotype 1
- G5:
-
Genotype 5
- G6:
-
Genotype 6
- N:
-
Number
- NADH-1:
-
NADH dehydrogenase subunit 1
References
Saad MB, Magzoub M. Echinococcus granulosus infection in Tamboul. Sudan J Helminthol. 1986;60:299–300.
Omer RA, Dinkle A, Romig T, Mackenstedt U, Elnahas AA, Aradaib IE, Ahmed ME, Elmalik KH, Adam A. A molecular survey of cystic echinococcosis in Sudan. Vet Parasitol. 2010;169:340–6.
Wahlers K, Menezes CN, Wong ML, Zeyhle E, Ahmed ME, Ocaido M, Stijnis C, Romig T, Kern P, Grobusch MP. Cystic echinococcosis in sub-Saharan Africa. Lancet Infect Dis. 2012;12:871–80.
Ahmed ME, Eltom KH, Musa NO, Ali IA, Elamin FM, Grobusch MP, Aradaib IE. First report on circulation of Echinococcus ortleppi in the one humped camel (Camelus dromedaries), Sudan. BMC Vet Res. 2013;9:127.
Eckert J, Deplazes P. Biological, epidemiological, and clinical aspects of echinococcosis, a zoonosis of increasing concern. Am Soc Mic. 2004;17:107–35.
Torgerson PR. Economic effect of echinococcosis. Acta Trop. 2003;85:113–8.
Musa NO, Eltom K, Awad S, Gameel AA. Causes of condemnation of sheep carcasses in abattoirs in Khartoum. In: Tielkes E, editor. Tropentag book of abstracts: Tropentag 19–21 Sept. 2012. Göttingen: Cuvillier; 2012. p. 54.
Budke C, Deplazes P, Torgerson P. Global socioeconomic impact of cystic echinococcosis. Emerg Infect Dis. 2006;12:296–303.
Deplazes P, Rinaidi L, Rojas A, Harandi MF, Romig T, Antolova D, Jm S, Lahmar S, Cringol G, Magambo J, Thompson RCA, Jenkins EJ. Global distribution of alveolar and cystic echinococcosis. Adv Parasitol. 2017;95:315–493.
Thompson RC. The taxonomy, phylogeny and transmission of Echinococcus. Exp Parasitol. 2008;119:439–46.
Brunetti E, Kern P, Vuitton D. (writing panel for the WHO-IWGE). Expert consensus for the diagnosis and treatment of cystic and alveolar echinococcosis in humans. Acta Trop. 2010;114:1–16.
Sadjjadi S. Present situation of echinococcosis in the Middle East and Arabic North Africa. Parasitol Int. 2006;55:197–202.
Taha H. Genetic variations among Echinococcus granulosus isolates in Egypt using RAPD-PCR. Parasitol Res. 2012;111:1993–2000.
Zhang W, Zhang Z, Wu W, Shi B, Li J, Zhou X, et al. Epidemiology and control of echinococcosis in central Asia, with particular reference to the People's Republic of China. Acta Trop. 2014;141:235–43.
Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol. 1992;54:165–73.
Bowles J, Blair D, McManus DP. A molecular phylogeny of the genus Echinococcus. Parasitology. 1995;110:317–28.
Bowles J, McManus DP. Rapid discrimination of Echinococcus species and strains using a polymerase chain reaction-based RFLP method. Mol Biochem Parasitol. 1993;57:231–9.
Bowles J, McManus DP. NADH dehydrogenase 1 gene sequences compared for species and strains of the genus Echinococcus. Int J Parasitol. 1993;23:969–72.
Lavikainen A, Lehtinen MJ, Meri T, Hirvila-Koski V, Meri S. Molecular genetic characterization of the Fennoscandien cervid strain, a new genotypic group (G10) of Echinococcus granulosus. Parasitology. 2003;127:207–15.
McManus DP. The molecular epidemiology of Echinococcus granulosus and cystic hydatid disease. Trans R Soc Trop Med Hyg. 2002;96:51–157.
Elmahdi IE, Ali QM, Magzoub MM, Ibrahim AM, Saad MB, Romig T. Cystic echinococcosis of livestock and humans in central Sudan. Ann Trop Med Parasitol. 2004;98:473–9.
Ibrahim K, Romig T, Peter K, Omer RA. A molecular survey on cystic echinococcosis in Sinnar area, Blue Nile state (Sudan). Chinese Med J. 2011;124:2829–33.
Ahmed ME, Abdelrahim MI, Ahmed FM. Hydatid disease, a morbid drop needs awareness. Sudan Med J. 2011;47:4–8.
Nakao M, Li T, Han X, Ma X, Xiao N, Qiu J, Wang H, Yanagida T, Mamuti W, Wen H, Moro PL, Giraudoux P, Craig PS, Ito A. Genetic polymorphisms of Echinococcus tapeworms in China as determined by mitochondrial and nuclear DNA sequences. Int J Parasitol. 2010;40:7.
Thompson RC, Lymbery AJ, Constantine CC. Variation in Echinococcus: towards a taxonomic revision of the genus. Adv Parasitol. 1995;35:145–76.
Thompson RC, Lymbery AJ. The nature, extent and significance ofvariation within the genus Echinococcus. Adv Parasitol. 1988;27:209–58.
Abushhewa M, Abushhiwa M, Nolan M, Jex A, Campbell B, Jabbar A, et al. Genetic classification of Echinococcus granulosus cysts from humans, cattle and camels in Libya using mutation scanning-based analysis of mitochondrial loci. Mol Cell Probes. 2010;24:346–51.
Eisa AM, Mustfa AA, Soliman KN. Preliminary report on cysticercosis and hydatidosis in the southern Sudan. Sud J Vet Sc Anim Husb. 1962;3:97–102.
El Khawat SE, Eisa AM, Slepnev NK, Saad MB. Hydatidosis of domestic animals in the central region of the Sudan. Bull Anim Hlth Prod Afr. 1979;27:249–51.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/. Nt Nucl Acids Symp Ser. 1999;41:95–8.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48.
Alvarez, Rojas CA, Romig T, Lightowlers. M.W. Echinococcus granulosus sensu lato genotypes infecting humans—review of current knowledge. Int J Parasitol. 2014;44:9–18.
Acknowledgments
We thank the Veterinary Officers at Al-kadarou slaughterhouse for many assistance to this study. The authors would also like to thank Mr. Abdalla M. Fadlemoula for technical assistance. The findings and conclusions in this report are those derived by the authors and do not necessarily represent the views of the funding source. The authors would also like to thank Professor M.O. Khedir for English editing of the manuscript.
Funding
This study has no available funding.
Availability of data and materials
The data and materials of this study were linked to the Dryad repository via TreeBASE. The link to TreeBASE search ID is provided below: https://doi.org/105061/dryad.5qr6t.
Author information
Authors and Affiliations
Contributions
MEA help with collection of hydatid cyst samples, extracted the DNA, optimized the polymerase chain reaction-based detection assay, editing of sequences and helped with the manuscript writting; BS collected hydatid cyst samples, edited and analyzed the sequence data; MPG designed the experiment and helped with preparation of the final manuscript; IEA designed the experiment, helped with collection of hydatid cyst samples and prepared the final manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the Institutional Research Board (IRB), Deanship of Scientific Research, Al-Neelain University, Khartoum, Sudan. Hydatid cysts were collected from slaughtered cattle during post-mortem inspection by qualified veterinary officers at Al-kadarou slaughter house, Khartoum North, Sudan. Formal consent and permission for research use of hydatid cysts were obtained from both the university and abattoir veterinarians. In this study, no experiment was conducted on live animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests. All authors have read and approved the final version of this manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ahmed, M.E., Salim, B., Grobusch, M.P. et al. First molecular characterization of Echinococcus granulosus (sensu stricto) genotype 1 among cattle in Sudan. BMC Vet Res 14, 36 (2018). https://doi.org/10.1186/s12917-018-1348-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-018-1348-9