Abstract
Background
Magnetic resonance imaging (MRI) is the modality of choice for diagnosing and monitoring muscular tissue pathologies and bone marrow alterations in the context of lower back pain, neuromuscular diseases and osteoporosis. Chemical shift encoding-based water-fat MRI allows for reliable determination of proton density fat fraction (PDFF) of the muscle and bone marrow. Prior to quantitative data extraction, segmentation of the examined structures is needed. Performed manually, the segmentation process is time consuming and therefore limiting the clinical applicability. Thus, the development of automated segmentation algorithms is an ongoing research focus.
Construction and content
This database provides ground truth data which may help to develop and test automatic lumbar muscle and vertebra segmentation algorithms. Lumbar muscle groups and vertebral bodies (L1 to L5) were manually segmented in chemical shift encoding-based water-fat MRI and made publically available in the database MyoSegmenTUM. The database consists of water, fat and PDFF images with corresponding segmentation masks for lumbar muscle groups (right/left erector spinae and psoas muscles, respectively) and lumbar vertebral bodies 1–5 of 54 healthy Caucasian subjects. The database is freely accessible online at https://osf.io/3j54b/?view_only=f5089274d4a449cda2fef1d2df0ecc56.
Conclusion
A development and testing of segmentation algorithms based on this database may allow the use of quantitative MRI in clinical routine.
Similar content being viewed by others
Background
Morphology-based magnetic resonance imaging (MRI) is clinically used to identify and monitor pathological changes of the spine in many diseases (e.g. osteoporosis or lower back pain) [1,2,3]. Quantitative MR including magnetic resonance spectroscopy and chemical shift encoding-based water-fat MRI allows for reliable determination of muscle and bone marrow fat composition [4, 5]. It can be used for the quantitative assessment of muscular tissue composition in neuromuscular disorders and their longitudinal monitoring [6, 7]. It was reported that intramuscular fat is increased in patients with neuromuscular diseases and lower back pain [3, 7,8,9,10]. Bone marrow malignancies, osteoporosis and distinction of the fractures are also common fields of application for quantitative MRI [11,12,13]. Compared to quantitative MRI, morphology-based MRI with the subsequent qualitative analysis and generation of semiquantitative parameters is limited in this context considering interrater objectivity and experience-based follow up evaluation [14].
Proton density fat fraction (PDFF) can be calculated based on chemical shift encoding-based water-fat MRI [15, 16]. However, up to now a manually segmentation process is needed to extract PDFF values from the lumbar muscles and the vertebral bone marrow. This is time consuming and therefore limiting the clinical applicability. Clinicians as well as scientists would strongly benefit from automated segmentation methods. Despite some recent developments in (semi-) automated segmentation techniques in different imaging disciplines their transfer into clinical routine is still challenging [17,18,19]. Previous work reported possibilities for muscle and vertebrae segmentation in MR images [20,21,22,23]. These algorithms require large datasets to test their performance and to further develop them. However, the public availability of manually segmented datasets of lumbar muscle groups and vertebrae as ground truth are limited.
Therefore, the purpose of this work was to provide a ground truth database with quantitative chemical shift encoding-based water-fat MR images and segmentations of the lumbar muscle groups (right/left erector spinae and psoas muscles, respectively) and the lumbar vertebral bodies (L1 to L5).
Construction and content
Subjects
The database contains 54 MRI datasets of healthy, Caucasian volunteers (15 males, 39 female, age: 51.6 ± 16.7 years).
MR imaging
Fifty-four healthy volunteers underwent MRI on a 3 T system (Ingenia, Philips Healthcare, Best, Netherlands) using a whole-body coil, the built-in 12-channel posterior coil and a 16-channel anterior coil. Subjects were positioned head-first in a supine position. Two different sequences were used for imaging the lumbar muscles and vertebral bodies. The conducted scanning protocol included dedicated scanning parameters, which are shown in Table 1.
The acquired data were analyzed using the mDIXON fat quantification method provided by the manufacturer. A complex-based water-fat decomposition was performed using a single T2* correction and a pre-calibrated fat spectrum accounting for the presence of the multiple peaks in the fat spectrum [24]. A seven-peak fat spectrum model was employed. The imaging-based proton density fat fraction (PDFF) map was computed as the ratio of the fat signal over the sum of fat and water signals, described previously by Schlaeger et al. [25, 26].
Axial and sagittal water, fat and PDFF images were stored as separate datasets for each subject as a *.dcm file.
MR image segmentation
The subsequent segmentation of the computed PDFF was performed manually using the open access image viewer software MITK (Medical Imaging Interaction Toolkit, Heidelberg, Germany). All segmentations were performed by a board certified radiologist.
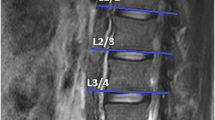
The vertebral bodies L1 to L5 were segmented in the sagittal PDFF maps excluding the posterior elements (Fig. 1).
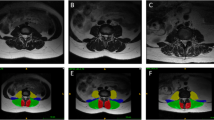
The axial PDFF maps were used to separately segment the erector spinae and the psoas muscle on both sides from the cranial part of L2 to caudal part of L5. Figure 2 exemplarily shows the depicted ROI in the corresponding muscles.
PDFF of each vertebral body (L1 to L5) and muscle group (right/left erector spinae and psoas muscle, respectively) were extracted. The segmentations of each vertebral body and muscle group is available as a binary mask. Each mask was stored as a separate *.mha file.
Utility and discussion
Database availability
The database is available online at https://osf.io/3j54b/?view_only=f5089274d4a449cda2fef1d2df0ecc56. Axial and sagittal water, fat and PDFF images are deposited as separate datasets for each subject as a *.dcm file. The segmentation masks of each vertebra (L1 to L5) and muscle group were deposited as *.mha files.
Subjects’ characteristics
Datasets of subjects and corresponding segmentation masks are labeled with the same subject ID (1 to 54). Masks were labeled as L1 to L5 or muscle group (right erector spinae muscle, left erector spinae muscle, right psoas muscle, and left psoas muscle, respectively).
Subject characteristics (sex, age [years], weight [kg], height [m] and body mass index (BMI) [kg/m2]) and PDFF values [%] of the vertebral bone marrow (L1 to L5) and the lumbar muscle groups (right/left erector spinae and psoas muscle, respectively) are listed in Table 2. Segmentation of the paraspinal muscles and the lumbar vertebrae (L1 to L5) amounted 50 min and 1:40 h for each subject, respectively.
Discussion
Recent and future trends of imaging automatization are and surely will be influencing many aspects of radiological diagnostics now and in the future. Subspecialties as neuroradiology, breast imaging, musculoskeletal radiology and oncological imaging are affected regarding (semi-) automated initial diagnostic finding, disease monitoring and report classification [18, 27,28,29]. Our aim with offering a database for manually segmented lumbar muscles and vertebral bodies (L1 to L5) in MR images of 54 healthy volunteers is to provide ground truth data which supports testing and refining newly developed computer vision or machine learning algorithms for automatic lumbar muscle and spine segmentation. The present database offers access to water, fat and PDFF maps.
With the extractable volume and PDFF values for the psoas and erector spinae muscles and the vertebral bodies L1 to L5, we elucidate the use of quantitative MRI in spine and muscle imaging and offer a ground truth dataset for automatic algorithm testing purposes. Figure 1 exemplarily illustrates the PDFF map of the vertebral bodies L1 to L5 and Fig. 2 shows a PDFF map for the respective muscle groups. The presented data is in line with the studies of Schlaeger et al. and Baum et al. and could help to develop more efficient ways of segmenting musculoskeletal structures [25, 30]. This is necessary to enable scientists and clinicians to perform data processing and analysis in an economic way. Additionally, the described methodical aspects are of great importance in order to being able to transfer automatized quantitative MRI into clinical routine by promoting computational segmenting and analyzing PDFF maps. In a further step the engineering of a fully automatic diagnosis finding due to pattern based deep learning could be possible and highly beneficial to diagnostic accuracy, interreader reliability and longitudinal comparability (Fig. 3) [31]. As recently shown in skin cancer diagnostics huge amounts of datasets are needed to constitute an algorithm quality that can outperform a specialist’s evaluation [28]. The amount of data generated with each qualitative respectively quantitative MRI scan delineates information which needs to be assessed carefully and precisely. This sets the high standards to every algorithm competing with specialists’ knowledge and skills. Complex disease entities like neuromuscular disorders, osteoporosis or lower back pain can be represented in different anatomical structures from very subtle bony or muscular alterations to striking pathologic imaging findings in cases of fractures or muscle atrophy [2, 5, 32, 33].
This database should be considered as one contribution to an ongoing process of rationalization and automatization. The limitation of the present database is the inclusion of healthy subjects only. Thus, this database has to be extended by diseased subjects in the future.
Conclusion
We present the database MyoSegmenTUM Spine with manually segmented lumbar muscle groups and vertebral bodies in MR images of 54 healthy volunteers together with corresponding manual segmentation masks. The data can be used as a training and test datasets for the development of automatic lumbar muscle and spine segmentation algorithms. These algorithms are highly needed to promote and accelerate the wide spread clinical implementation of quantitative muscle MRI for diagnosis of muscle and vertebral pathologies.
Abbreviations
- A/P:
-
Anterior/posterior
- FOV:
-
Field of view
- L/R:
-
Left/right
- L1, L2 etc.:
-
Lumbar vertebrae 1, lumbar vertebrae 2 etc.
- MRI:
-
Magnetic Resonance Imaging
- PDFF:
-
Proton density fat fraction
- TE:
-
Echo time
- TR:
-
Repetition time
References
Nouh MR, Eid AF. Magnetic resonance imaging of the spinal marrow: basic understanding of the normal marrow pattern and its variant. World J Radiol. 2015;7(12):448–58. https://doi.org/10.4329/wjr.v7.i12.448 Epub 12/01/2016. PubMed PMID: 26753060; PubMed Central PMCID: PMCPMC4697119.
Link TM. Osteoporosis imaging: state of the art and advanced imaging. Radiology. 2012;263(1):3–17. https://doi.org/10.1148/radiol.12110462 Epub 23/03/2012. PubMed PMID: 22438439; PubMed Central PMCID: PMCPMC3309802.
Wan Q, Lin C, Li X, Zeng W, Ma C. MRI assessment of paraspinal muscles in patients with acute and chronic unilateral low back pain. Br J Radiol. 2015;88(1053):20140546. https://doi.org/10.1259/bjr.20140546 Epub 25/06/2015. PubMed PMID: 26105517; PubMed Central PMCID: PMCPMC4743557.
Karampinos DC, Ruschke S, Dieckmeyer M, Diefenbach M, Franz D, Gersing AS, et al. Quantitative MRI and spectroscopy of bone marrow. J Magn Reson Imaging. 2018;47(2):332–53. https://doi.org/10.1002/jmri.25769 Epub 02/06/2017. PubMed PMID: 28570033; PubMed Central PMCID: PMCPMC5811907.
Baum T, Cordes C, Dieckmeyer M, Ruschke S, Franz D, Hauner H, et al. MR-based assessment of body fat distribution and characteristics. Eur J Radiol. 2016;85(8):1512–8. https://doi.org/10.1016/j.ejrad.2016.02.013 Epub 26/02/2016. PubMed PMID: 26905521.
Carlier PG, Azzabou N, de Sousa PL, Hicks A, Boisserie JM, Amadon A, et al. Skeletal muscle quantitative nuclear magnetic resonance imaging follow-up of adult Pompe patients. J Inherit Metab Dis. 2015;38(3):565–72. https://doi.org/10.1007/s10545-015-9825-9 Epub 10/03/2015. PubMed PMID: 25749708; PubMed Central PMCID: PMCPMC4432102.
Forbes SC, Willcocks RJ, Triplett WT, Rooney WD, Lott DJ, Wang DJ, et al. Magnetic resonance imaging and spectroscopy assessment of lower extremity skeletal muscles in boys with Duchenne muscular dystrophy: a multicenter cross sectional study. PLoS One. 2014;9(9):e106435. https://doi.org/10.1371/journal.pone.0106435 Epub 10/09/2014. PubMed PMID: 25203313; PubMed Central PMCID: PMCPMC4159278.
Kim HK, Laor T, Horn PS, Racadio JM, Wong B, Dardzinski BJ. T2 mapping in Duchenne muscular dystrophy: distribution of disease activity and correlation with clinical assessments. Radiology. 2010;255(3):899–908. https://doi.org/10.1148/radiol.10091547 Epub 27/05/2010. PubMed PMID: 20501727.
Pezolato A, de Vasconcelos EE, Defino HL, Nogueira-Barbosa MH. Fat infiltration in the lumbar multifidus and erector spinae muscles in subjects with sway-back posture. Eur Spine J. 2012;21(11):2158–64. https://doi.org/10.1007/s00586-012-2286-z Epub 03/04/2012. PubMed PMID: 22465969; PubMed Central PMCID: PMCPMC3481104.
Hodges P, Holm AK, Hansson T, Holm S. Rapid atrophy of the lumbar multifidus follows experimental disc or nerve root injury. Spine (Phila Pa 1976). 2006;31(25):2926–33. https://doi.org/10.1097/01.brs.0000248453.51165.0b Epub 02/12/2006. PubMed PMID: 17139223.
Jung HS, Jee WH, McCauley TR, Ha KY, Choi KH. Discrimination of metastatic from acute osteoporotic compression spinal fractures with MR imaging. Radiographics. 2003;23(1):179–87. https://doi.org/10.1148/rg.231025043 Epub 21/01/2003. PubMed PMID: 12533652.
Shah LM, Hanrahan CJ. MRI of spinal bone marrow: part I, techniques and normal age-related appearances. AJR Am J Roentgenol. 2011;197(6):1298–308. https://doi.org/10.2214/AJR.11.7005 Epub 24/11/2011. PubMed PMID: 22109283.
Silva JR Jr, Hayashi D, Yonenaga T, Fukuda K, Genant HK, Lin C, et al. MRI of bone marrow abnormalities in hematological malignancies. Diagn Interv Radiol. 2013;19(5):393–9. https://doi.org/10.5152/dir.2013.067 Epub 12/06/2013. PubMed PMID: 23748035.
Mercuri E, Pichiecchio A, Allsop J, Messina S, Pane M, Muntoni F. Muscle MRI in inherited neuromuscular disorders: past, present, and future. J Magn Reson Imaging. 2007;25(2):433–40. https://doi.org/10.1002/jmri.20804 Epub 30/01/2007. PubMed PMID: 17260395.
Hernando D, Sharma SD, Aliyari Ghasabeh M, Alvis BD, Arora SS, Hamilton G, et al. Multisite, multivendor validation of the accuracy and reproducibility of proton-density fat-fraction quantification at 1.5T and 3T using a fat-water phantom. Magn Reson Med. 2017;77(4):1516–24. https://doi.org/10.1002/mrm.26228 Epub 16/04/2016. PubMed PMID: 27080068; PubMed Central PMCID: PMCPMC4835219.
Reeder SB, Hu HH, Sirlin CB. Proton density fat-fraction: a standardized MR-based biomarker of tissue fat concentration. J Magn Reson Imaging. 2012;36(5):1011–4. https://doi.org/10.1002/jmri.23741 Epub 11/07/2012. PubMed PMID: 22777847; PubMed Central PMCID: PMCPMC4779595.
Baudin PY, Azzabou N, Carlier PG, Paragios N. Prior knowledge, random walks and human skeletal muscle segmentation. Med Image Comput Comput Assist Interv. 2012;15(Pt 1):569–76 Epub 05/01/2013. PubMed PMID: 23285597.
Huber T, Alber G, Bette S, Boeckh-Behrens T, Gempt J, Ringel F, et al. Reliability of semi-automated segmentations in glioblastoma. Clin Neuroradiol. 2017;27(2):153–61. https://doi.org/10.1007/s00062-015-0471-2 Epub 23/10/2015. PubMed PMID: 26490369.
Valcarcel AM, Linn KA, Vandekar SN, Satterthwaite TD, Muschelli J, Calabresi PA, et al. MIMoSA: An Automated Method for Intermodal Segmentation Analysis of Multiple Sclerosis Brain Lesions. J Neuroimaging. 2018. https://doi.org/10.1111/jon.12506 Epub 09/03/2018. PubMed PMID: 29516669.
Brunner G, Nambi V, Yang E, Kumar A, Virani SS, Kougias P, et al. Automatic quantification of muscle volumes in magnetic resonance imaging scans of the lower extremities. Magn Reson Imaging. 2011;29(8):1065–75. https://doi.org/10.1016/j.mri.2011.02.033 Epub 23/08/2011. PubMed PMID: 21855242.
Karlsson A, Rosander J, Romu T, Tallberg J, Gronqvist A, Borga M, et al. Automatic and quantitative assessment of regional muscle volume by multi-atlas segmentation using whole-body water-fat MRI. J Magn Reson Imaging. 2015;41(6):1558–69. https://doi.org/10.1002/jmri.24726 Epub 12/08/2014. PubMed PMID: 25111561.
Chu C, Belavy DL, Armbrecht G, Bansmann M, Felsenberg D, Zheng G. Fully automatic localization and segmentation of 3D vertebral bodies from CT/MR images via a learning-based method. PLoS One. 2015;10(11):e0143327. https://doi.org/10.1371/journal.pone.0143327 Epub 26/11/2015. PubMed PMID: 26599505; PubMed Central PMCID: PMCPMC4658120.
Gaonkar B, Xia Y, Villaroman DS, Ko A, Attiah M, Beckett JS, et al. Multi-parameter ensemble learning for automated vertebral body segmentation in heterogeneously acquired clinical MR images. IEEE J Transl Eng Health Med. 2017;5:1800412. https://doi.org/10.1109/JTEHM.2017.2717982 Epub 12/10/2017. PubMed PMID: 29018631; PubMed Central PMCID: PMCPMC5515511.
Ren J, Dimitrov I, Sherry AD, Malloy CR. Composition of adipose tissue and marrow fat in humans by 1H NMR at 7 Tesla. J Lipid Res. 2008;49(9):2055–62. https://doi.org/10.1194/jlr.D800010-JLR200 Epub 30/05/2008. PubMed PMID: 18509197; PubMed Central PMCID: PMCPMC2515528.
Schlaeger S, Freitag F, Klupp E, Dieckmeyer M, Weidlich D, Inhuber S, et al. Thigh muscle segmentation of chemical shift encoding-based water-fat magnetic resonance images: the reference database MyoSegmenTUM. PLoS One. 2018;13(6):e0198200. https://doi.org/10.1371/journal.pone.0198200 Epub 08/06/2018. PubMed PMID: 29879128.
Schlaeger S, Inhuber S, Rohrmeier A, Dieckmeyer M, Freitag F, Klupp E, et al. Association of paraspinal muscle water-fat MRI-based measurements with isometric strength measurements. Eur Radiol. 2019;29(2):599–608. https://doi.org/10.1007/s00330-018-5631-8 Epub 18/07/2018. PubMed PMID: 30014202.
Bahl M, Barzilay R, Yedidia AB, Locascio NJ, Yu L, Lehman CD. High-risk breast lesions: a machine learning model to predict pathologic upgrade and reduce unnecessary surgical excision. Radiology. 2018;286(3):810–8. https://doi.org/10.1148/radiol.2017170549 Epub 19/10/2017. PubMed PMID: 29039725.
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–8. https://doi.org/10.1038/nature21056 Epub 25/01/2017. PubMed PMID: 28117445.
Chen MC, Ball RL, Yang L, Moradzadeh N, Chapman BE, Larson DB, et al. Deep learning to classify radiology free-text reports. Radiology. 2018;286(3):845–52. https://doi.org/10.1148/radiol.2017171115 Epub 15/11/2017. PubMed PMID: 29135365.
Baum T, Yap SP, Dieckmeyer M, Ruschke S, Eggers H, Kooijman H, et al. Assessment of whole spine vertebral bone marrow fat using chemical shift-encoding based water-fat MRI. J Magn Reson Imaging. 2015;42(4):1018–23. https://doi.org/10.1002/jmri.24854 Epub 03/02/2015. PubMed PMID: 25639780.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. https://doi.org/10.1038/nature14539 Epub 29/05/2015. PubMed PMID: 26017442.
Sambrook P, Cooper C. Osteoporosis. Lancet. 2006;367(9527):2010–8. https://doi.org/10.1016/S0140-6736(06)68891-0 Epub 20/06/2006. PubMed PMID: 16782492.
Karampinos DC, Baum T, Nardo L, Alizai H, Yu H, Carballido-Gamio J, et al. Characterization of the regional distribution of skeletal muscle adipose tissue in type 2 diabetes using chemical shift-based water/fat separation. J Magn Reson Imaging. 2012;35(4):899–907. https://doi.org/10.1002/jmri.23512 Epub 01/12/2011. PubMed PMID: 22127958; PubMed Central PMCID: PMCPMC3292710.
Acknowledgements
Not Applicable.
Funding
The present work was supported by the European Research Council (grant agreement No 677661 – ProFatMRI and grant agreement No 637164 – iBack), TUM Faculty of Medicine KKF grant H01 and Philips Healthcare. The funding bodies had no influence in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The database presented in this study is freely available online at https://osf.io/3j54b/?view_only=f5089274d4a449cda2fef1d2df0ecc56.
Author information
Authors and Affiliations
Contributions
Conception and design: TB, JSK, DCK. Analysis and interpretation of data: EB, TB. Drafting of the article: EB, TB. Critical revision of the article for important intellectual content: EB; AR, SS, MD, MND, DW, JS, CZ, EK, EJR, JSK, DCK, TB. Final approval of the article: EB; AR, SS, MD, MND, DW, JS, CZ, EK, EJR, JSK, DCK, TB. Collection and assembly of data: AR, SS, MD, MND, DW. Obtaining funding: TB, JSK, DCK. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the local institutional Committee for Human Research (Ethikkommission der Fakultät für Medizin, Technische Universität München, Munich, Germany). All subjects gave written informed consent before participation in the study.
Consent for publication
Written informed consent for publication was obtained from all patients included in this study.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Burian, E., Rohrmeier, A., Schlaeger, S. et al. Lumbar muscle and vertebral bodies segmentation of chemical shift encoding-based water-fat MRI: the reference database MyoSegmenTUM spine. BMC Musculoskelet Disord 20, 152 (2019). https://doi.org/10.1186/s12891-019-2528-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12891-019-2528-x