Abstract
Background
Low lung function has been associated with increased body mass index (BMI). The aim of this study was to investigate whether the effect of BMI on lung function is mediated by DNA methylation.
Methods
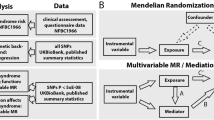
We used individual data from 285,495 participants in four population-based cohorts: the European Community Respiratory Health Survey, the Northern Finland Birth Cohort 1966, the Swiss Study on Air Pollution and Lung Disease in Adults, and the UK Biobank. We carried out Mendelian randomisation (MR) analyses in two steps using a two-sample approach with SNPs as instrumental variables (IVs) in each step. In step 1 MR, we estimated the causal effect of BMI on peripheral blood DNA methylation (measured at genome-wide level) using 95 BMI-associated SNPs as IVs. In step 2 MR, we estimated the causal effect of DNA methylation on FEV1, FVC, and FEV1/FVC using two SNPs acting as methQTLs occurring close (in cis) to CpGs identified in the first step. These analyses were conducted after exclusion of weak IVs (F statistic < 10) and MR estimates were derived using the Wald ratio, with standard error from the delta method. Individuals whose data were used in step 1 were not included in step 2.
Results
In step 1, we found that BMI might have a small causal effect on DNA methylation levels (less than 1% change in methylation per 1 kg/m2 increase in BMI) at two CpGs (cg09046979 and cg12580248). In step 2, we found no evidence of a causal effect of DNA methylation at cg09046979 on lung function. We could not estimate the causal effect of DNA methylation at cg12580248 on lung function as we could not find publicly available data on the association of this CpG with SNPs.
Conclusions
To our knowledge, this is the first paper to report the use of a two-step MR approach to assess the role of DNA methylation in mediating the effect of a non-genetic factor on lung function. Our findings do not support a mediating effect of DNA methylation in the association of lung function with BMI.
Similar content being viewed by others
Background
There are several cross-sectional studies showing lower forced respiratory volumes (FEV1 and FVC) in those who are overweight and obese. Evidence on the association between obesity and FEV1/FVC is less clear [1]. Longitudinal studies also suggest that an increase in body mass index (BMI) is associated with increased lung function decline. An increase in BMI among overweight and obese people has been associated with greater than average decline of FEV1 and FVC [2, 3]. There is always a risk that such associations may be confounded, but a large Mendelian randomisation (MR) study has shown that increasing BMI leads to the decline in both FEV1 and FVC [4], suggesting that this association is causal.
The underlying mechanisms for this association are unclear with some hypothesising that it reflects pulmonary biological responses to obesity and its related pro-inflammatory status, and others suggesting it reflects thoracic compression (i.e., a mechanical effect) [5, 6]. As both differences in BMI and lung function have been associated with DNA methylation levels [7,8,9,10], we hypothesised that association of lung function with BMI is in part mediated by DNA methylation.
A new method using MR to investigate the mediating effect of DNA methylation on the association of risk factors and health outcomes has recently been described [11, 12]. This two-step epigenetic MR approach relies on the use of genetic variants potentially controlling DNA methylation. In the first step, a single nucleotide polymorphism (SNP), or group of SNPs, that proxies for the risk factor of interest (here BMI) is used to assess the causal relationship between the risk factor and DNA methylation. If this association is confirmed, in the second step, a SNP that proxies for methylation levels at the site modified by the risk factor is used to interrogate the causal relationship between DNA methylation and the main outcome (here lung function). We applied this technique to investigate the mediating role of DNA methylation in the association of lung function with BMI in European cohorts, after using a conventional non-MR approach.
Methods
Conventional (non-MR) analysis
We assessed the cross-sectional association of lung function (FEV1, FVC, FEV1/FVC) with BMI in participants of the European Community Respiratory Health Survey (ECRHS, n = 470), the Northern Finland Birth Cohort 1966 (NFBC1966, n = 681), and the Swiss Study on Air Pollution and Lung Disease in Adults (SAPALDIA, n = 962) (see Supplementary File 1 for details) who also had information on peripheral blood DNA methylation. This analysis was carried out using linear regression models adjusted for centre, sex, age, height, sex-age interaction, sex-height interaction, educational level, smoking status, and pack-years of smoking. We estimated the association of each lung function parameter with BMI for each cohort, and then combined them in a random effects meta-analysis.
Two-step MR
We carried out MR analyses in two steps using a two-sample approach for summary data with SNPs as instruments in each step. To assess the strength of each SNP as instrument, we calculated the F statistic [13], and to avoid bias due to use of weak instruments, we included in the MR analyses only SNPs with an F statistic equal to or greater than 10 [14]. Individual-level data used in these analyses come from four cohorts: ECRHS, NFBC1966, SAPALDIA, and UK Biobank (see Supplementary File 1 for details). Analyses were conducted using R v.3.3.2 [15].
First-step MR: examining the causal effect of BMI on DNA methylation
SNP-BMI association estimates
A recent published genome-wide association meta-analysis of 125 studies on 339,224 participants reported the association of BMI with 97 SNPs (accounting for an estimated 2.7% of the variability of BMI in the population) [16]. We extracted their effect estimates and standard errors and used these SNPs as instruments in the first step MR (Fig. 1: GX1).
SNP-DNA methylation estimates
We created a weighted genetic risk score (wGRS) for BMI from the 97 SNPs for 470 participants from the ECRHS and 681 participants from the NFBC1966. The wGRS was the sum of the products of the effect allele dosage at each of the 97 SNPs and their corresponding beta coefficients [16]. As a screening stage we used linear regression to measure the effect of the wGRS on DNA methylation (Table 1). CpGs associated with BMI-wGRS at P < 10− 7 were then examined to assess their association with individual SNPs (Fig. 1: GY1) contributing to the wGRS. Replication of the identified associations was sought in the SAPALDIA cohort (n = 906) (Table 1). These analyses were adjusted for ancestry principal components. Participants included in this analysis are those included in the conventional (non-MR) analysis, except for 56 participants from SAPALDIA for whom there were no allele dosage data.
BMI-DNA methylation estimates: MR analysis
To estimate the causal effect of BMI on DNA methylation levels, we derived MR estimates for each of the 95 SNPs with an F statistic equal to or greater than 10 (Table E1 in Supplementary File 1), using the Wald estimator (ratio of the genotype-outcome regression coefficient to the genotype-exposure regression coefficient), with standard error derived using the delta method [17]. The individual MR estimates were combined using inverse-variance weighted (IVW) fixed-effect meta-analysis [18]. To investigate the robustness of the MR findings to pleiotropy, we used the following methods: 1) IVW random-effects meta-analysis [19]; 2) Egger regression with penalized weights [20]; and 3) weighted median analysis [21].
Second-step MR: examining the causal effect of DNA methylation on lung function
DNA methylation-cis-SNP association estimates
Using the publicly available mQTL database (http://mqtldb.org; accessed on 1 December 2017), which contains the associations of peripheral blood DNA methylation with SNPs as observed in the ALSPAC-ARIES project, we identified SNPs associated with the CpGs discovered in step 1 (p < 10− 7) and located within 1 Mb either side of the CpG (cis-SNPs) [22] (Fig. 1: GX2). We selected independent cis-SNPs, after linkage disequilibrium (LD) clumping (‘clump_data’ function from R package ‘TwoSampleMR’), as the instruments for the DNA methylation of interest. The regression coefficient and standard error for the cis-SNP-methylation association were used in the MR analysis.
Cis-SNP-lung function association estimates
We regressed lung function parameters (FEV1, FVC, FEV1/FVC) on the cis-SNPs (Fig. 1: GY2) in the ECRHS (n = 773), NFBC1966 (n = 4501), SAPALDIA (n = 2303), and UK Biobank (n = 275,861) cohorts (Table 1). The analysis was adjusted for ancestry principal components and did not include data from participants included in step one. As a sex difference in the association of lung function with BMI has been reported [2], FEV1, FVC and FEV1/FVC were adjusted for sex.
DNA methylation-lung function estimates: MR analysis
To estimate the causal effect of DNA methylation on lung function, we derived MR estimates for each SNP with an F statistic equal to or greater than 10 using the Wald estimator, with standard error derived using the delta method [17].
Results
A description of the BMI and lung function parameters of the participants in the several cohorts included in this analysis is presented in Table 1. On average, BMI was similar across cohorts and lung volumes were higher in NFBC1966 and SAPALDIA.
Conventional (non-Mendelian randomisation) analysis
A higher BMI was associated with lower FEV1 (beta coefficient = − 0.009, 95% CI − 0.019 to 0.0) and FVC (beta coefficient = − 0.19, 95% CI − 0.03 to − 0.009) among participants of ECRHS, NFBC and SAPALDIA who had DNA methylation. The FEV1/FVC ratio was positively associated with BMI (beta coefficient = 0.0013, 95% CI 0.0005 to 0.002) (Fig. 2).
First-step MR: examining the causal effect of BMI on DNA methylation
The genotype-BMI association estimates for the 97 SNPs used as instrumental variables for BMI are presented in Table E1 (see Supplementary File 1). In the screening stage, the 97-SNP wGRS was associated with two CpGs, one in SBK1 (cg09046979) and one in NPIPB11 (cg12580248) (Table E2 in Supplementary File 1). MR estimates, from IVW fixed effect meta-analysis, for the effect of BMI on these two CpGs are presented in Table 2. A 1-unit increase in BMI was responsible for less than 1% change in methylation at either of the CpGs. The effect of BMI on cg09046979 was statistically significant in ECRHS and NFBC1966, but not in SAPALDIA. The effect of BMI on cg12580248 was not statistically significant in ECRHS and NFBC1966 and could not be assessed in SAPALDIA as a probe for this CpG is not present in the methylation chip used in SAPALDIA. Results from the IVW random effects meta-analysis, Egger regression and weighted median analysis were consistent with those of the IVW fixed effect meta-analysis (Fig. 3).
Second-step MR: examining the causal effect of DNA methylation on lung function
Using the mQTLdb and after LD clumping, we found that methylation at CpG cg09046979 had been associated with two independent SNPs (rs9938394 and rs9939450) occurring within 1 Mb either side of the probe (cis-SNPs). We could not find publicly available information from independent cohorts on genotype-DNA methylation associations where the Infinium MethylationEPIC BeadChip was used to measure levels of methylation. The genotype-DNA methylation association estimates for the two SNPs, which were used as instrumental variables for DNA methylation, are presented in Table E3 (see Supplementary File 1).
MR estimates for the effect of DNA methylation, at cg09046979, on FEV1, FVC and the FEV1/FVC ratio are presented in Table 3. There was no evidence of an association between methylation levels at this site and lung function.
Discussion
The findings of this 2-step epigenetic MR study suggest a small causal effect of BMI on DNA methylation at one or two CpGs, but also suggest that these are unlikely to exert a causal effect on lung function.
As this is a multicentre study across several countries, confounding due to population stratification is possible. However, most study participants were of European descent and ancestry principal components were included in the analyses. There is always concern within an MR study that pleiotropy (when an SNP affects several phenotypes related to the outcome [23], in this case, DNA methylation in the first step MR and lung function in the second step MR) may exist and there was some evidence of heterogeneity (I2 of 41% for cg09046979 and 43% for cg12580248) suggestive of pleiotropy. However, we used methods that are robust to pleiotropy (i.e. IVW random effects, Egger regression, and weighted median) and found results to be consistent with those of the IVW fixed effect analysis. The sample size of the first step MR was limited by the relatively small number of ECRHS, NFBC1966 and SAPALDIA participants with available data on DNA methylation, which may have reduced the chances of identifying causal associations of BMI with DNA methylation. Despite the very large sample size in the second step MR, our capacity to fully explore our findings for one of the CpGs was limited by the lack of information on associations between SNPs and CpG methylation assessed using the EPIC (850 K) chip from independent studies. All studies that utilise peripheral blood DNA methylation data to explore associations of lifestyle and environmental factors with organ specific abnormalities are limited by the lack of consistent clear evidence that DNA methylation in peripheral blood reflects well what is going on in the relevant disease tissue. Although some concordance in DNA methylation levels between blood and lung tissue has been reported [24] as well as for BMI related blood methylation with that in adipose tissue [25], some argue this is unlikely to be common [26]. As variation in DNA methylation levels across the epigenome is often tissue-specific [27], we cannot for sure say that the association of lung function with BMI is not mediated by DNA methylation within lung tissue.
Conclusion
In conclusion, our findings do not support a mediating effect of peripheral blood DNA methylation in the association of lung function with BMI.
Availability of data and materials
Access to individual level data is restricted. However, data requests for sound research proposals will be addressed. For ECRHS data, contact DLJ (d.jarvis@imperial.ac.uk). For NFBC1966 data, contact MRJ (m.jarvelin@imperial.ac.uk). For SAPALDIA data, contact NMPH (nicole.probst@swisstph.ch). For UK Biobank data, request should be made through www.ukbiobank.ac.uk.
Abbreviations
- BMI:
-
Body mass index
- CpG:
-
5′-C-phosphate-G-3
- DNA:
-
Deoxyribonucleic acid
- ECRHS:
-
European community respiratory health survey
- FEV1 :
-
Forced expiratory volume in one second
- FVC:
-
Forced vital capacity
- IQR:
-
Interquartile range
- IV:
-
Instrumental variable
- IVW:
-
Inverse-variance weighted
- LD:
-
Linkage disequilibrium
- methQTL:
-
Methylation quantitative trait locus
- MR:
-
Mendelian randomisation
- NFBC:
-
Northern Finland birth cohort
- SAPALDIA:
-
Swiss study on air pollution and lung disease in adults
- SNP:
-
Single nucleotide polymorphism
- wGRS:
-
Weighted genetic risk score
References
Forno E, Han YY, Mullen J, Celedon JC. Overweight, obesity, and lung function in children and adults-a meta-analysis. J Allergy Clin Immunol Pract. 2018;6:570–81 e510.
Carey IM, Cook DG, Strachan DP. The effects of adiposity and weight change on forced expiratory volume decline in a longitudinal study of adults. Int J Obes Relat Metab Disord. 1999;23:979–85.
Thyagarajan B, Jacobs DR Jr, Apostol GG, Smith LJ, Jensen RL, Crapo RO, Barr RG, Lewis CE, Williams OD. Longitudinal association of body mass index with lung function: the CARDIA study. Respir Res. 2008;9:31.
Skaaby T, Taylor AE, Thuesen BH, Jacobsen RK, Friedrich N, Mollehave LT, Hansen S, Larsen SC, Volker U, Nauck M, Volzke H, Hansen T, Pedersen O, Jorgensen T, Paternoster L, Munafo M, Grarup N, Linneberg A. Estimating the causal effect of body mass index on hay fever, asthma and lung function using Mendelian randomization. Allergy. 2018;73:153–64.
Baffi CW, Wood L, Winnica D, Strollo PJ Jr, Gladwin MT, Que LG, Holguin F. Metabolic syndrome and the lung. Chest. 2016;149:1525–34.
Salome CM, King GG, Berend N. Physiology of obesity and effects on lung function. J Appl Physiol. 2010;108:206–11.
Machin M, Amaral AF, Wielscher M, Rezwan FI, Imboden M, Jarvelin MR, Adcock IM, Probst-Hensch N, Holloway JW, Jarvis DL, study A. Systematic review of lung function and COPD with peripheral blood DNA methylation in population based studies. 2017.
Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, Tsai PC, Ried JS, Zhang WH, Yang YW, Tan SL, Fiorito G, Franke L, Guarrera S, Kasela S, Kriebel J, Richmond RC, Adamo M, Afzal U, Ala-Korpela M, Albetti B, Ammerpohl O, Apperley JF, Beekman M, Bertazzi PA, Black SL, Blancher C, Bonder MJ, Brosch M, Carstensen-Kirberg M, de AJM C, de Lusignan S, Dehghan A, Elkalaawy M, Fischer K, Franco OH, Gaunt TR, Hampe J, Hashemi M, Isaacs A, Jenkinson A, Jha S, Kato N, Krogh V, Laffan M, Meisinger C, Meitinger T, Mok ZY, Motta V, Ng HK, Nikolakopoulou Z, Nteliopoulos G, Panico S, Pervjakova N, Prokisch H, Rathmann W, Roden M, Rota F, Rozario MA, Sandling JK, Schafmayer C, Schramm K, Siebert R, Slagboom PE, Soininen P, Stolk L, Strauch K, Tai ES, Tarantini L, Thorand B, Tigchelaar EF, Tumino R, Uitterlinden AG, van Duijn C, van JBJ M, Vineis P, Wickremasinghe AR, Wijmenga C, Yang TP, Yuan W, Zhernakova A, Batterham RL, Smith GD, Deloukas P, Heijmans BT, Herder C, Hofman A, Lindgren CM, Milani L, van der Harst P, Peters A, Illig T, Relton CL, Waldenberger M, Jarvelin MR, Bollati V, Soong R, Spector TD, Scott J, McCarthy MI, Elliott P, Bell JT, Matullo G, Gieger C, Kooner JS, Grallert H, Chambers JC. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81.
Lee MK, Hong Y, Kim SY, Kim WJ, London SJ. Epigenome-wide association study of chronic obstructive pulmonary disease and lung function in Koreans. Epigenomics. 2017;9:971–84.
Imboden M, Wielscher M, Rezwan FI, AFS A, Schaffner E, Jeong A, Beckmeyer-Borowko A, Harris SE, Starr JM, Deary IJ, Flexeder C, Waldenberger M, Peters A, Schulz H, Chen S, Sunny SK, WJJ K, Jiang Y, Erhart G, Kronenberg F, Arathimos R, Sharp GC, Henderson AJ, Fu Y, Piirila P, Pietilainen KH, Ollikainen M, Johansson A, Gyllensten U, de Vries M, van der Plaat DA, de Jong K, Boezen HM, Hall IP, Tobin MD, Jarvelin MR, Holloway JW, Jarvis D, Probst-Hensch NM. Epigenome-wide association study of lung function level and its change. Eur Respir J. 2019;54:1900457.
Caramaschi D, Sharp GC, Nohr EA, Berryman K, Lewis SJ, Davey Smith G, Relton CL. Exploring a causal role of DNA methylation in the relationship between maternal vitamin B12 during pregnancy and child's IQ at age 8, cognitive performance and educational attainment: a two-step Mendelian randomization study. Hum Mol Genet. 2017;26:3001–13.
Relton CL, Davey SG. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41:161–76.
Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ, Smith GD, Sterne JAC. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012;21:223–42.
Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Smith GD. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–63.
R Core Team. R. A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2016.
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Felix R, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan JA, Magi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Zhao JH, Zhao W, Chen J, Fehrmann R, Hedman AK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham L, Buyske S, Demirkan A, Deng GH, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Leach IM, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stancakova A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Settee J, Van Vliet-Ostaptchouk JV, Wang ZM, Yengo L, Zhang WH, Isaacs A, Albrecht E, Arnlov J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Bluher M, Bohringer S, Bonnycastle LL, Bottcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Chen YD, Clarke R, Daw EW, de AJM C, Delgado G, Dimitriou M, ASF D, Eklund N, Estrada K, Eury E, Folkersen L, Fraser RM, Garcia ME, Geller F, Giedraitis V, Gigante B, Go AS, Golay A, Goodall AH, Gordon SD, Gorski M, Grabe HJ, Grallert H, Grammer TB, Grassler J, Gronberg H, Groves CJ, Gusto G, Haessler J, Hall P, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hengstenberg C, Holmen O, Hottenga JJ, James AL, Jeff JM, Johansson A, Jolley J, Juliusdottir T, Kinnunen L, Koenig W, Koskenvuo M, Kratzer W, Laitinen J, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindstrom J, Lo KS, Lobbens S, Lorbeer R, Lu YC, Mach F, PKE M, Mahajan A, McArdle WL, McLachlan S, Menni C, Merger S, Mihailov E, Milani L, Moayyeri A, Monda KL, Morken MA, Mulas A, Muller G, Muller-Nurasyid M, Musk AW, Nagaraja R, Nothen MM, Nolte IM, Pilz S, Rayner NW, Renstrom F, Rettig R, Ried JS, Ripke S, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Scott WR, Seufferlein T, Shi JX, Smith AV, Smolonska J, Stanton AV, Steinthorsdottir V, Stirrups K, Stringham HM, Sundstrom J, Swertz MA, Swift AJ, Syvanen AC, Tan ST, Tayo BO, Thorand B, Thorleifsson G, Tyrer JP, Uh HW, Vandenput L, Verhulst FC, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Warren HR, Waterworth D, Weedon MN, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wrightl AF, Zhang QY, Brennan EP, Choi M, Dastani Z, Drong AW, Eriksson P, Franco-Cereceda A, Gadin JR, Gharavi AG, Goddard ME, Handsaker RE, Huang JY, Karpe F, Kathiresan S, Keildson S, Kiryluk K, Kubo M, Lee JY, Liang L, Lifton RP, Ma BS, SA MC, AJ MK, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Okada Y, JRB P, Dorajoo R, Reinmaa E, Salem RM, Sandholm N, Scott RA, Stolk L, Takahashi A, Tanaka T, van ’t Hooft FM, AAE V, Westra HJ, Zheng W, Zondervan KT, Heath AC, Arveiler D, SJL B, Beilby J, Bergman RN, Blangero J, Bovet P, Campbell H, Caulfield MJ, Cesana G, Chakravarti A, Chasman DI, Chines PS, Collins FS, Crawford DC, Cupples LA, Cusi D, Danesh J, de Faire U, den Ruijter HM, Dominiczak AF, Erbel R, Erdmann J, Eriksson JG, Farrall M, Felix SB, Ferrannini E, Ferrieres J, Ford I, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, Gejman PV, Gieger C, Gottesman O, Gudnason V, Gyllensten U, Hall AS, Harris TB, Hattersley AT, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Homuth G, Hovingh GK, Humphries SE, Hunt SC, Hypponen E, Illig T, Jacobs KB, Jarvelin MR, Jockel KH, Johansen B, Jousilahti P, Jukema JW, Jula AM, Kaprio J, JJP K, Keinanen-Kiukaanniemi SM, Kiemeney LA, Knekt P, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Le Marchand L, Lehtimaki T, Lyssenko V, Mannisto S, Marette A, Matise TC, McKenzie CA, McKnight B, Moll FL, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, PAF M, Pasterkamp G, Peden JF, Peters A, Postma DS, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Rioux JD, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramines J, Sarzynski MA, Schunkert H, PEH S, Sever P, Shuldiner AR, Sinisalo J, Stolk RP, Strauch K, Tonjes A, Tregouet DA, Tremblay A, Tremoli E, Virtamo J, Vohl MC, Volker U, Waeber G, Willemsen G, Witteman JC, Zillikens MC, Adair LS, Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bornstein SR, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de PIW B, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hui JN, Hunter DJ, Hveem K, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu YM, Martin NG, Marz W, Melbve M, Metspalu A, Moebus S, Munroe PB, Njolstad I, Oostra BA, CNA P, Pedersen NL, Perola M, Perusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sattar N, Schadt EE, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Thorsteinsdottir UR, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Walker M, Wallaschofski H, Wareham NJ, Watkins H, Weir DR, Wichmann HE, Wilson JF, Zanen P, Borecki IB, Deloukas P, Fox CS, Heid IM, O'Connell JR, Strachan DP, Stefansson K, van Duijri CM, Abecasis GR, Franke L, Frayling TM, McCarthy MI, Visscher PM, Scherag A, Willer CJ, Boehnke M, Mohlke KL, Lindgren CM, Beckmann JS, Barroso I, North KE, Ingelsson E, Hirschhorn JN, RJF L, Speliotes EK, Study LC, Consortium A, Grp A-BW, Consortium CD, Consortium C, GLGC, ICBP, Investigators M, Consortium M, Consortium M, Consortium P, Consortium R, Consortium G, Consortium IE. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–U401.
Thompson JR, Minelli C, Del Greco MF. Mendelian randomization using public data from genetic consortia. Int J Biostat. 2016;12.
Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65.
Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med. 2017.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. 2015;44:512–25.
Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14.
Gaunt TR, Shihab HA, Hemani G, Min JL, Woodward G, Lyttleton O, Zheng J, Duggirala A, McArdle WL, Ho K, Ring SM, Evans DM, Davey Smith G, Relton CL. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016;17:61.
Stearns FW. One hundred years of pleiotropy: a retrospective. Genetics. 2010;186:767–73.
Stueve TR, Li WQ, Shi J, Marconett CN, Zhang T, Yang C, Mullen D, Yan C, Wheeler W, Hua X, Zhou B, Borok Z, Caporaso NE, Pesatori AC, Duan J, Laird-Offringa IA, Landi MT. Epigenome-wide analysis of DNA methylation in lung tissue shows concordance with blood studies and identifies tobacco smoke-inducible enhancers. Hum Mol Genet. 2017;26:3014–27.
Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, Tsai PC, Ried JS, Zhang W, Yang Y, Tan S, Fiorito G, Franke L, Guarrera S, Kasela S, Kriebel J, Richmond RC, Adamo M, Afzal U, Ala-Korpela M, Albetti B, Ammerpohl O, Apperley JF, Beekman M, Bertazzi PA, Black SL, Blancher C, Bonder MJ, Brosch M, Carstensen-Kirberg M, de Craen AJ, de Lusignan S, Dehghan A, Elkalaawy M, Fischer K, Franco OH, Gaunt TR, Hampe J, Hashemi M, Isaacs A, Jenkinson A, Jha S, Kato N, Krogh V, Laffan M, Meisinger C, Meitinger T, Mok ZY, Motta V, Ng HK, Nikolakopoulou Z, Nteliopoulos G, Panico S, Pervjakova N, Prokisch H, Rathmann W, Roden M, Rota F, Rozario MA, Sandling JK, Schafmayer C, Schramm K, Siebert R, Slagboom PE, Soininen P, Stolk L, Strauch K, Tai ES, Tarantini L, Thorand B, Tigchelaar EF, Tumino R, Uitterlinden AG, van Duijn C, van Meurs JB, Vineis P, Wickremasinghe AR, Wijmenga C, Yang TP, Yuan W, Zhernakova A, Batterham RL, Smith GD, Deloukas P, Heijmans BT, Herder C, Hofman A, Lindgren CM, Milani L, van der Harst P, Peters A, Illig T, Relton CL, Waldenberger M, Jarvelin MR, Bollati V, Soong R, Spector TD, Scott J, MI MC, Elliott P, Bell JT, Matullo G, Gieger C, Kooner JS, Grallert H, Chambers JC. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–6.
Daley D, Shey K, Akhabir L, Saferali A, Mah SM, Sandford A, Kobor MS, Pare P. Comparison of methylation profiles in human blood and lung tissue identifies tissue specific CpG methylation sites. Respirology. 2014;19:17.
De Bustos C, Ramos E, Young JM, Tran RK, Menzel U, Langford CF, Eichler EE, Hsu L, Henikoff S, Dumanski JP, Trask BJ. Tissue-specific variation in DNA methylation levels along human chromosome 1. Epigenetics Chromatin. 2009;2:7.
Acknowledgements
We would like to thank the ECRHS, NFBC1966, SAPALDIA and UK Biobank participants, field workers and researchers for their time and cooperation. In addition, we thank the late Professor Paula Rantakallio (launch of NFBC1966), Ms. Outi Tornwall and Ms. Minttu Jussila (DNA biobanking), the late Academian of Science Leena Peltonen, and Mr. James Potts.
Funding
This work was conducted within the Ageing Lungs in European Cohorts (ALEC) project and received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 633212. The funders of this study had no role in study design, data analysis and interpretation of results, or writing of the manuscript.
The ECRHS was supported by a contract from the European Commission (018996), Fondo de Investigación Sanitaria (91/0016–060-05/E, 92/0319, 93/0393, 97/0035–01, 99/0034–01 and 99/0034–02), Hospital General de Albacete, Hospital General Ramón Jiménez, Consejería de Sanidad del Principado de Asturias, CIRIT (1997SGR 00079, 1999SGR 00241), and Servicio Andaluz de Salud, SEPAR, Public Health Service (R01 HL62633–01), RCESP (C03/09), Red RESPIRA (C03/011), Basque Health Department, Swiss National Science Foundation, Swiss Federal Office for Education and Science, Swiss National Accident Insurance Fund (SUVA), GSF-National Research Centre for Environment and Health, Deutsche Forschungsgemeinschaft (DFG) (FR 1526/1–1, MA 711/4–1), Programme Hospitalier de Recherche Clinique-DRC de Grenoble 2000 no. 2610, Ministry of Health, Direction de la Recherche Clinique, Ministere de l’Emploi et de la Solidarite, Direction Generale de la Sante, CHU de Grenoble, Comite des Maladies Respiratoires de l’Isere. UCB-Pharma (France), Aventis (France), Glaxo France. Estonian Science Foundation, and Asthma UK (formerly known as National Asthma Campaign UK).
The NFBC resource has been supported by grants from the Academy of Finland (project grants 104781, 120315, 129269, 1114194, 24300796, Center of Excellence in Complex Disease Genetics and SALVE), University Hospital Oulu, Biocenter, University of Oulu, Finland (75617), NHLBI grant 5R01HL087679–02 (1RL1MH083268–01), NIH/NIMH (5R01MH63706:02), ENGAGE project and grant agreement HEALTH-F4–2007-201413, EU FP7 EurHEALTHAgeing − 277849, the Medical Research Council, UK (G0500539, G0600705, G1002319, PrevMetSyn/SALVE) and the MRC, Centenary Early Career Award. H2020 DynaHEALTH (European Union’s Horizon 2020 research and innovation programme under grant agreement No 633595); Exposomic, Genomic and Epigenomic Approach to Prediction of Metabolic and Cardiorespiratory function and Ill-Health (EGEA), Academy of Finland, Grant No 285547; ALEC Study (funded by the European Union’s Horizon 2020 Research and Innovation programme under grant agreement No. 633212); H2020 / Marie Skłodowska-Curie Actions, CAPICE (Marie Curie Grant agreement Number 721567); National Public Health Institute, Biomedicum Helsinki, Finland.
The SAPALDIA study was funded by the Swiss National Science Foundation (grants no 33CS30–148470/1&2, 33CSCO-134276/1, 33CSCO-108796, 324730_135673, 3247BO-104283, 3247BO-104288, 3247BO-104284, 3247–065896, 3100–059302, 3200–052720, 3200–042532, 4026–028099, PMPDP3_129021/1, PMPDP3_141671/1), the Federal Office for the Environment, the Federal Office of Public Health, the Federal Office of Roads and Transport, the canton’s government of Aargau, Basel-Stadt, Basel-Land, Geneva, Luzern, Ticino, Valais, and Zürich, the Swiss Lung League, the canton’s Lung League of Basel Stadt/ Basel Landschaft, Geneva, Ticino, Valais, Graubünden and Zurich, Stiftung ehemals Bündner Heilstätten, SUVA, Freiwillige Akademische Gesellschaft, UBS Wealth Foundation, Talecris Biotherapeutics GmbH, Abbott Diagnostics, European Commission 018996 (GABRIEL), Wellcome Trust WT 084703MA, Exposomics EC FP7 grant (Grant agreement No: 308610).
Author information
Authors and Affiliations
Consortia
Contributions
AFSA, CM, JWH, MRJ, NMPH and DJ designed the study. AFSA, MI, MW, FIR, AJ and ES analysed the data. AFSA, MW, FIR, CM, JGA, GPP, JA, AJ, ES, ABB, JWH, MRJ, NMPH and DLJ interpreted and discussed the results, and read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The ECRHS study was approved by the local ethics committees in each region: Reykjavík, Iceland (The National Bioethics Committee, Reykjavík, Iceland); Umea, Uppsala, Gothenburg, Sweden (Regional Ethical Committee in Uppsala, Sweden); Erfurt, Hamburg, Germany (Ethic Committee of the Bavarian State Chamber of Physicians, Germany); Norwich, UK (Norwich District Ethics Committee); Ipswich, UK (Ipswich–East Suffolk Local Research Ethics Committee); Grenoble, France (Ethics committee Paris Bichat-Claude Bernard); Barcelona, Spain (Comité Ético de Investigación Clínica del Instituto Municipal de Asistencia Sanitaria, Barcelona, Spain); Albacete, Spain (Comité de Ética e Investigación de Complejo Hospitalario de Albacete, Spain); Oviedo, Spain (Comité Ético de Investigación Clínica Regional, Hospital Universitario Central de Asturias, Oviedo, Spain); Galdakao, Spain (Comité Ético de Investigación del Hospital de Galdakao, Spain); and Basel, Switzerland (Swiss Academy of Medical Sciences and the ethics committee of Basel). All participants provided informed written consent.
The NFBC1966 study was approved by the ethics committees in Oulu (Finland) and Oxford (UK) universities. All participants provided informed written consent.
The SAPALDIA study was approved by the Overall Regional Ethics Commission for Clinical Medicine (Swiss Academy of Medical Sciences) and by the respective cantonal ethical committee for each survey. All participants provided informed written consent.
The UK Biobank was approved by the U.K. National Research Ethics Service Committee NorthWest – Haydock. All participants provided informed written consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Supplementary File 1.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Amaral, A.F.S., Imboden, M., Wielscher, M. et al. Role of DNA methylation in the association of lung function with body mass index: a two-step epigenetic Mendelian randomisation study. BMC Pulm Med 20, 171 (2020). https://doi.org/10.1186/s12890-020-01212-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12890-020-01212-9