Abstract
Background
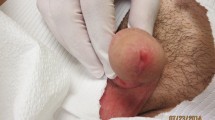
Leishmaniasis is a parasitic infection endemic in more than ninety countries of the world. The cutaneous leishmaniasis (CL) is a most common form of leishmaniasis and it remains to be a major public health issue in Saudi Arabia. This study was undertaken to investigate the Leishmania species responsible for CL infection in different provinces of Qassim, Saudi Arabia.
Methods
Skin biopsies were obtained from CL patients and DNA was extracted using the Magna pure system. Leishmania species were identified by highly specific/sensitive quantitative and qualitative PCR.
Results
Out of total 206 CL biopsies, 49.5% biopsies were found to be positive for Leishmania major (L. major), 28.6% biopsies were positive for Leishmania tropica (L. tropica), 3.9% were found to be positive for Leishmania infantum/donovani (L. infantum/donovani). Not only have these, all tested CL biopsies showed negative test for Leishmania mexicana (L. mexicana) and Leishmania viannia (L. viannia).
Conclusions
This is the first comprehensive study that shows the majority of CL in Qassim was caused by L. major and L. tropica. To the best of our knowledge, this is the very first report that shows the occurrence of L. infantum/donovani in Saudi Arabia. This requires higher alert to the Ministry of Health of Saudi Arabia to take proactive actions in preventing the onset of L. major, L. tropica, L. infantum and L. donovani infections.
Similar content being viewed by others
Background
Leishmaniasis is a parasitic infection endemic in more than ninety countries of the world and the cutaneous leishmaniasis (CL) is a most common form of this infection caused by phlebotomine sand fly [1]. The World Health Organization reported in 2016 that about 15 million individuals have leishmaniasis and more than 360 million individuals are breathing in those regions which are prone for this infection and this infection causes ~ 70,000 deaths per year [1, 2]. It is now well documented that CL is caused by more than 22 different species of the genus Leishmania (L) but their prevalence varies from region to region [3, 4]. Identification of specific L. species is important for the prescription of appropriate therapy [5]. Treatment of CL patients without identification of L. species cause harmful effects to the patients as the attribution of the relative importance of specific L. species in humans are reported [6]. In general, diagnosis is still based on clinical symptoms, microscopic parasitic detection and tissue culturing of promastigotes. However in cases with promastigotes culture, additional efforts should also be needed such as biochemical and serological analysis for further characterization of parasites [7]. These additional efforts for the determination of L. species are time taken and are not sensitive, not accurate and sometime give wrong information [8]. Recently, several PCR based detection of L. species were developed, which are rapid, sensitive and accurate and now become a powerful approach to determine the L. species types at all levels of detection such as genus, complex, and species [9, 10]. Recently, Abuzaid et al. have extensively reviewed the prevalence of CL in Saudi Arabia. They noticed that CL remains an unsolved public health issue of the country [10]. Although, CL is endemic in all over the regions of Saudi Arabia but the majority of patients are continuously reported in Riyadh, Hassa, Aseer, Hail, Madinah, Taif and also in Qassim [10,11,12,13]. Despite of all taking care by the Ministry of Health, Saudi Arabia but CL remains to be a major health issue of the country, which may be due to urbanization and huge population immigration [10]. As the information on the diversity of L. species in different provinces of Saudi Arabia is poor, therefore the current study was aimed to identify the L. species in different provinces of Qassim, Saudi Arabia using highly specific and sensitive PCR-based approach.
Methods
Patients recruitment, biopsies collection and DNA extraction
The study was carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki as revised in Tokyo 2004) for humans and the study protocol was approved by the National Plan for Science, Technology and Innovation of Saudi Arabia (NSTIP/KACST # 11-MED1068–09). With the Institutional Review Board (IRB) approval, study subjects were recruited through the dermatology outpatient clinics of Qassim region of Saudi Arabia. Patients were diagnosed after careful clinical examination based on clinical presentation and microscopy and were classified as cutaneous leishmaniasis as described previously [14]. Demographic details of the studied subjects are shown in Table 1. A total of 206 biopsies were taken from cutaneous lesions and DNA samples were extracted from all biopsies by MagNaA pure DNA extraction Pure LC DNA Isolation Kit (cat. # 03186229001, Roche Applied Science, Mannheim, Germany) using MagNA Pure LC Automated Instrument according to the manufacturer’s instruction (Roche) as described previously [15, 16]. The absorbance of DNA solution was monitored at 260 nm and 280 nm to ascertain its concentration and purity by using Perkin Elmer UV-Spectrophotometer (L7110223-D, Lambda XLS, Germany) as described previously [17, 18].
Real time PCR
Quantitative real-time PCR amplification was carried out for the detection of L. major, L. viannia, L. mexicana and L. infantum/donovani by the Light cycler PCR system (Roche Diagnostics, Mannheim, Germany). By the use of CLUSTAL program in Lasergene software (Madison, WI), the amplicons were aligned and nucleotide differences among the various Leishmania complexes were determined as described previously [19]. Using primer express software (PE Biosystems, Foster City, CA), appropriate probes and flanking primers were designed to specifically identify each Leishmania complex. The following primers/probe combination for each complex was designed: L. major complex: F-5’TTCTGCTCCGTCGGTGTAGA3’, R-5’GCTTTCGATTGGCTACGACAA3’; and Lmaj-probe 5′-CCTGTCAGGAATTCCACAAA-3′; L. infantum/donovani complex: F-5’CCAGATGCCGACCAAAGC3’, R-5’CGCGCACGTGATGGATAAC3’ and Lid-probe 5′-ATCGGCAGGTTCT-3′; L. mexicana complex: F-5’CCAGTCCCAGAACACAAACATG3’, R-5’CCTATCGACCAACACAGAAAAGG3’ and Lm probe 5′-ATGCCGAACTCCCGAA-3′; L. viannia complex: F-5’CAACAAAATGCTTCGCAACAG3’, R-5’CGCAACGCCTTCATGGA3’ and LV-probe 5′-CGACGGGATATTGTTTGACTT-3′. PCR amplification with specific primer for L. species was performed in the same ways as described previously [19,20,21] with some modifications. Briefly, typical profile times used were initial step, 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 15 s. and annealing/extension at 60 °C for 30 s, followed by 4.0 °C up to 24 h. The reactions were conducted in a 20 μl and set up in a 96-well optical reaction plate. The reactants mix are containing 1x fast start master mix (Integrated DNA Technologies, USA), 3.0 pg. to 100 ng of DNA template, 1 μM of each primer (Integrated DNA Technologies, USA). The presence of amplified products (a positive result) was determined by melting curves and values were recorded for estimation the L. species. All samples were run again with conventional PCR assays to determine L. tropica species.
Qualitative PCR
The qualitative PCR for L. tropica was carried out by using the following primers sequence F-5’TCGTCTGATTCAAAGTTCTC3’, R-5’CACACGCGCACACCGCGATC3’ designed from Leishmania tropica GTG1 sequence (GenBank: AY826393.1) as described previously [22]. The Go Taq® Green Mix (cat. # M7123, Promega Corporation, WI, USA) was used according the instructions provided by the manufacturer (Promega, WI, USA). Briefly, the reaction mixture (50 μL) contained 1X Go Taq® Green Master Mix, 1 μM of each primers (Integrated DNA Technologies, USA), and 100–300 ng of the extracted DNA. The amplification was performed by MWG Biotech Inc. Primus 96 Thermal Cycler, 220 VAC (Mfr# TC-9600-226 V; Item # EW-93941-05) using an initial heating of 95 °C for 2 min. 40 cycles were applied with three stages: denaturation (95 °C for 30 s), annealing (55 for 30 s) and extension (72 °C for 1 min). The products of PCR were loaded to 2% agarose gel and electrophoresed at 10–15 V, followed by staining with 0.1 μg/mL ethyl bromide. The presence of bands was then detected by MacroVue UV-20 transilluminator (Hoefer, Inc., MA, USA).
Positive and negative controls samples
Positive control DNA samples were extracted from the cultures of standard WHO strains of L. major MHOM/SU/1973/5ASKH, L. tropica MHOM/SU/1974/SAF-K27, L. infantum MHOM/TN/1980/IPT1, L. donovani MHOM/IN/1980/DD8, L. mexicana MHOM/BZ/1982/BEL21, L. (viannia) braziliens MHOM/BR/1975/M2903. Negative control DNA samples were extracted from the clinical samples of different dermatological disorders.
Results
All skin biopsies were collected from cutaneous leishmaniasis (CL) patients, which were attending various dermatological clinics in different provinces of Qassim, Saudi Arabia. Sites of the study are shown in the map of Qassim province of Saudi Arabia (Fig. 1). The majority of the patients were from Buraidah (63%) and Unayzah (19%) and the rest (18%) were Ar Rass, Bukariya, Uglat Asugour, etc. In an attempt to investigate the different types of L. species in Qassim region, we used quantitative and qualitative PCR for identification of different L. species using specific sets of primers for L. major, L. infantum/donovani complex, L. mexicana, L. viannia, and L. tropica. Specificity and validity of each quantitative and qualitative PCR assays were tested by the standard WHO. stains. As shown in Table 2, there was a concordance in the characterization of L. species by both the quantitative and qualitative PCR assays. Moreover, the results obtained from PCR assays were further validated by using several negative control samples listed in Table 2. Furthermore, no amplification was detected with any of the PCR assays tested for the negative control DNA samples, whereas the presence of extracted DNA was confirmed by β-actin (Table 2). Amplification plots of positive samples for L. major, and L. infantum/donavani complex are presented in Fig. 2. These amplification plots of positive samples for L. major, and L. infantum/donavani complex were comparable with their respective positive controls. Importantly, all tested patients samples were found to be negative for L. viannia and L. maxicana. Negative controls and samples negative for specific L. species were not detected. Whereas, samples positive for L. tropica samples were characterized by agarose gel electrophoresis and the details are given in Fig. 3. Specifically, out of total 206 CL biopsies, 102 was found to be positive for L. major, 08 was positive for L. infantum/donovani complex, and 59 CL biopsies were found to be positive for L. tropica. We have also calculated the percent prevalence of these Leishmania species among the studied patients. The L. major and L. tropica are the most abundant types found in 49.5 and 28.6%, respectively, whereas L. infantum/donovani, was found in 3.9% of the studied subjects (Fig. 4). The complete data of the studied patients with the prevalence of L. species types in Qassim are summarized in Table 3.
(a) Map of Saudi Arabia showing Qassim region in yellow. (b) Map of Qassim region of Saudi Arabia showing sites of the study. The maps shown were taken from http://en.wikipedia.org/wiki/File:Saudi_Arabia_location_map.svg
Real time PCR amplification plots of positive samples for (a) Leishmania major complex, (b) Leishmania infantum/donavani complex, (c) Leishmania viannia complex, (d) Leishmania maxicana complex. Positive control for L. major was from strain MHOM/SU/1973/5ASKH, positive control for L. infantum/donavani complex was from strain MHOM/TN/1980/IPT1, positive control for L. mexicana was from strain MHOM/BZ/1982/BEL21, positive control for L. viannia was from L. braziliens strain MHOM/BR/1975/M2903. Negative controls and samples negative of specific L. species were not detected
Agarose gel electrophoresis for Leishmania tropica. Lane 1: molecular markers (cat. # 474, Biolabs); lane 2: PCR product of L. tropica strain MHOM/SU/1974/SAF-K27 (used as positive control); lane 3: PCR product of L. major strain MHOM/SU/1973/5ASKH (used as negative control); lane 4: PCR product of β-actin DNA (used as endogenous control); lane 5–12: PCR product from skin biopsies of cutaneous leishmaniasis cases
Discussion
This is the first comprehensive report that shows the majority CL in central region of Saudi Arabia was caused by L. major and L. tropica. Apply the PCR-based identification of specific L. species, this report also demonstrated the occurrence of L. infantum/donovani for the first time in Saudi Arabia. Based on the endemic nature of CL in gulf countries including Saudi Arabia and also unidentified L. species [10, 23], we have selected five Leishmania species namely L. major, L. infantum/donovani complex, L. mexicana, L. viannia, and L. tropica for the detection of CL infection. It is now well established that the PCR based identification of leishmaniasis infection has now been proved to be a better method as compared with other diagnostic methods in terms of specificity and accuracy [8, 9, 24, 25]. Quantitative real time PCR is an excellent diagnostic way which uses fluorescent labeling for the continuous watching of amplicon generation throughout the amplification process [8, 9, 26]. The most important advantages of quantitative real time PCR are that it is highly sensitive, accurate and extremely faster as compared to other available techniques for the detection of a target Leishmania species [8, 9, 19, 26]. Studies have shown that diagnosis of Leishmania species through quantitative real time PCR is 78 and 76% more sensitive than culture and expert microscopy, respectively [19, 24,25,26]. Therefore, we used quantitative real time PCR for all studied L. species except for L. tropica. In case of L. tropica and L. major, similar repeats in their sequences were reported at their flanking sequences and microsatellite, while designing of the PCR primer pairs. Furthermore, the previous studies also revealed that the microsatellites repeats were likely to be located on at least eight different chromosomes [22, 27]. For these reasons, the analysis of L. tropica was done through qualitative PCR analysis by using the well-designed GTG1 sequence (Gene bank: AY826393.1), which was well established for L. tropica only [22]. During the testing of all designed primers for PCR assays on positive and negative standard DNA samples, our results also showed that the primers on GTG1 sequence showed positive results only for L. tropica, whereas other tested L. species including L. major showed negative results.
In this study, CL infected skin biopsies were used from those CL patients which were attending various dermatological clinics in different provinces of Qassim, Saudi Arabia. The majority of the patients for this study were from Buraidah (63%) and Unaizah (19%) and the rest (18%) were from other parts of Qassim region including Ar Rass, Bukariya, Uglat Asugour (Fig. 1). All patients were diagnosed as CL based on clinical presentation and microscopy as described previously [14]. Our high sensitive PCR based analysis shows the L. major and L. tropica are the most abundant types of L. species found in 49.5 and 28.6% of CL patients, respectively. In support of our results, previous studies on parasite identification proved that L. major and L. tropica are the main causative CL species in Saudi Arabia [8, 10, 28, 29]. L. major has also been identified as the main Leishmania species in Riyadh and Al-Hassa regions [10] and now appears to be randomly transmitted in all over Saudi Arabia including Qassim [10, 28,29,30]. The characterization of isoenzyme L. major zymodeme LON-4 has been identified that L. major is a typical parasite present in all over Saudi Arabia [10]. Whereas, L. tropica has been found to be a main L. species responsible for causing CL infection in Aseer and Al-Bahah provinces which are in southwest part of Saudi Arabia [10, 28, 29]. In view of these and together with the occurrence of Phlebotomus papatasi, a L. major vector in many provinces of the country [10], it is now well established that L. major is responsible for causing CL infection in most of the provinces of the country, while L. tropica is only convicted in the southwestern zone [2]. However, recently it was demonstrated that L. tropica is also dispersed in other parts of the country other than southwestern area [10, 28,29,30]. As CL is well known to be an endemic disease not only in Saudi Arabia but also in other gulf countries and the main causative species were repeated found to be L. major and L. tropica in these countries [10, 23]. Addition of this study in different provinces of Qassim region has now been confirmed that the main causative Lieshmania species of cutaneous leishmaniasis in this region are L. major and L. tropica.
Anthroponotic transmission is not only the characteristic of L. tropica, but also for L. donovani complex, which is usually found in the Indian subcontinent including India, Bangladesh, Nepal, Sudan, Ethiopia, Kenya and Somalia [3, 31]. It is also reported that L. donovani is the cause of both anthroponotic and zoonotic characteristics in East Africa [3, 32]. On the other hand, L. infantum occurs in the Mediterranean, the Middle East, Afghanistan, Iran, Pakistan and Brazil, although sporadic cases have been reported in Central Asia, China, Mexico and Central and Latin America [31, 33]. Notably, immunosuppressed adults and children are at higher risk of clinical disorder due to L. infantum than immunocompetent adults [34]. Transmission of L. infantum infection is considered predominantly zoonotic, with domestic animals being the major reservoir [33, 35].
To the best of our knowledge, this is the first report that shows the occurrence of L. infantum/donovani complex in Saudi Arabia. Our data showed that 3.9% of studied leishmaniasis samples were found to be positive for L. infantum/donovani complex. The lower rate of occurrence of L. infantum/donovani complex in the studied region, suggested that this infection might be from outside of Saudi Arabia due to the huge population movement in the country especially from India, Bangladesh, Nepal, Sudan, Ethiopia, Kenya, Somalia, Afghanistan and Pakistan, where L. infantum/donovani complex is widely spread [3, 31, 32]. Not only have these, we have also tested the occurrence of L. mexicana and L. viannia in the studied patients samples, our test showed negative results for these L. species, strongly suggesting that the Qassim, the central region of Saudi Arabia is free of L. mexicana and L. viannia infection.
Conclusions
Cutaneous leishmaniasis is now considered to be a major dermatologic problem in Saudi Arabia with particular insight in the reported provinces. Using highly specific and sensitive PCR-based identification, this study determined that the majority cutaneous leishmaniasis patients in Qassim province of Saudi Arabia were caused by L. major and L. tropica. However, few leishmaniasis cases were found to be positive for L. infantum/donovani complex. This requires higher alert to the Ministry of Health of Saudi Arabia to take proactive actions in preventing the onset of L. major, L. tropica and L. infantum/donovani infections.
Abbreviations
- CL:
-
Cutaneous leishmaniasis
- KACST:
-
King Abdulaziz City for Science and Technology
- L. infantum/donovani:
-
Leishmania infantum/donovani
- L. major :
-
Leishmania major
- L. mexicana :
-
Leishmania mexicana
- L. tropica :
-
Leishmania tropica
- L. viannia:
-
Leishmania viannia
- NSTIP:
-
National Plan for Science, Technology and Innovation
References
Torres-Guerrero E, Quintanilla-Cedillo MR, Ruiz-Esmenjaud J, Arenas R. Leishmaniasis: a review. F1000Res. 2017;6:750.
World Health Organization: Weekly Epidemiological Record (WER)2016;91(22):285–296.
Georgiadou SP, Makaritsis KP, Dalekos GN. Leishmaniasis revisited: current aspects on epidemiology, diagnosis and treatment. J Transl Int Med. 2015;3(2):43–50.
Bailey F, Mondragon-Shem K, Hotez P, Ruiz-Postigo JA, Al-Salem W, Acosta-Serrano A, Molyneux DH. A new perspective on cutaneous leishmaniasis-implications for global prevalence and burden of disease estimates. PLoS Negl Trop Dis. 2017;11(8):e0005739.
Foulet F, Cosnes A, Dellion S, Blum L, Bachmeyer C, Raguin G, Tourte-Schaeffer C, Deniau M, Bretagne S. Leishmania major cutaneous leishmaniasis in HIV-positive patients does not spread to extralesional sites. Arch Dermatol. 2006;142(10):1368–9.
Wilson ME, Jeronimo SM, Pearson RD. Immunopathogenesis of infection with the visceralizing Leishmania species. Microb Pathog. 2005;38(4):147–60.
Eisenberger CL, Jaffe CL. Leishmania: identification of Old World species using a permissively primed intergenic polymorphic-polymerase chain reaction. Exp Parasitol. 1999;91(1):70–7.
Rasti S, Ghorbanzadeh B, Kheirandish F, Mousavi SG, Pirozmand A, Hooshyar H, Abani B. Comparison of molecular, microscopic, and culture methods for diagnosis of cutaneous Leishmaniasis. J Clin Lab Anal. 2016;30(5):610–5.
Mortarino M, Franceschi A, Mancianti F, Bazzocchi C, Genchi C, Bandi C. Quantitative PCR in the diagnosis of Leishmania. Parassitologia. 2004;46(1–2):163–7.
Abuzaid AA, Abdoon AM, Aldahan MA, Alzahrani AG, Alhakeem RF, Asiri AM, Alzahrani MH, Memish ZA. Cutaneous Leishmaniasis in Saudi Arabia: a comprehensive overview. Vector Borne Zoonotic Dis. 2017;17(10):673–84.
Elmekki MA, Elhassan MM, Ozbak HA, Qattan IT, Saleh SM, Alharbi AH. Epidemiological trends of cutaneous leishmaniasis in Al-Madinah Al-Munawarah province, western region of Saudi Arabia. J Glob Infect Dis. 2017;9(4):146–50.
Hawash YA, Ismail KA, Abdel-Wahab MM, Khalifa M. Diagnosis, treatment and clinical features of cutaneous Leishmaniasis in Saudi Arabia. Korean J Parasitol. 2018;56(3):229–36.
Jahan S, Al-Saigul AM, Nimir SE, Mustafa AS. Priorities for primary health care research in Qassim, Central Saudi Arabia. Saudi Med J. 2014;35(3):298–303.
Akilov OE, Khachemoune A, Hasan T. Clinical manifestations and classification of Old World cutaneous leishmaniasis. Int J Dermatol. 2007;46(2):132–42.
Alzolibani AA, Rasheed Z, Al Robaee AA. Acquired immunogenicity of DNA after modification with malondialdehyde in patients with alopecia areata. Scand J Clin Lab Invest. 2014;74(4):312–21.
Al-Shobaili HA, Ahmed AA, Rasheed Z. Recognition of oxidized albumin and thyroid antigens by psoriasis autoantibodies. A possible role of reactive-oxygen-species induced epitopes in chronic plaque psoriasis. Saudi Med J. 2015;36(12):1408–19.
Ahmad R, Rasheed Z, Kaushal E, Singh D, Ahsan H. Biochemical evaluation of human DNA-lysine Photoadduct treated with Peroxynitrite. Toxicol Mech Methods. 2008;18(7):589–95.
Al-Shobaili HA, Rasheed Z. Mitochondrial DNA acquires immunogenicity on exposure to nitrosative stress in patients with vitiligo. Hum Immunol. 2014;75(10):1053–61.
Wortmann G, Hochberg L, Houng HH, Sweeney C, Zapor M, Aronson N, Weina P, Ockenhouse CF. Rapid identification of Leishmania complexes by a real-time PCR assay. Am J Trop Med Hyg. 2005;73(6):999–1004.
Hossain F, Ghosh P, Khan MAA, Duthie MS, Vallur AC, Picone A, Howard RF, Reed SG, Mondal D. Real-time PCR in detection and quantitation of Leishmania donovani for the diagnosis of visceral Leishmaniasis patients and the monitoring of their response to treatment. PLoS One. 2017;12(9):e0185606.
Vallur AC, Duthie MS, Reinhart C, Tutterrow Y, Hamano S, Bhaskar KR, Coler RN, Mondal D, Reed SG. Biomarkers for intracellular pathogens: establishing tools as vaccine and therapeutic endpoints for visceral leishmaniasis. Clin Microbiol Infect. 2014;20(6):O374–83.
Schwenkenbecher JM, Fröhlich C, Gehre F, Schnur LF, Schönian G. Evolution and conservation of microsatellite markers for Leishmania tropica. Infect Genet Evol. 2004;4(2):99–105.
Salam N, Al-Shaqha WM, Azzi A. Leishmaniasis in the middle east: incidence and epidemiology. PLoS Negl Trop Dis. 2014;8(10):e3208.
Ertabaklar H, Ozlem Calıskan S, Boduc E, Ertug S. Comparison of direct microscopy, culture and polymerase chain reaction methods for the diagnosis of cutaneous leishmaniasis. Mikrobiyol Bul. 2015;49(1):77–84.
Akhoundi M, Downing T, Votýpka J, Kuhls K, Lukes J, Cannet A, Ravel C, Marty P, Delaunay P, Kasbari M, Granouillac B, Gradoni L, Sereno D. Leishmania infections: molecular targets and diagnosis. Mol Asp Med. 2017;57:1–29.
de Morais RC, da Costa Oliveira CN, de Albuquerque Sda C, Mendonça Trajano Silva LA, Pessoa-E-Silva R, Alves da Cruz HL, de Brito ME, de Paiva Cavalcanti M. Real-time PCR for Leishmania species identification: evaluation and comparison with classical techniques. Exp Parasitol. 2016;165:43–50.
Rossi V, Wincker P, Ravel C, Blaineau C, Pagés M, Bastien P. Structural organisation of microsatellite families in the Leishmania genome and polymorphisms at two (CA) n loci. Mol Biochem Parasitol. 1994;65(2):271–82.
Khan W, Zakai HA. Epidemiology, pathology and treatment of cutaneous leishmaniasis in taif region of Saudi Arabia. Iran J Parasitol. 2014;9(3):365–73.
El-Beshbishy HA, Al-Ali KH, El-Badry AA. Molecular characterization of cutaneous leishmaniasis in Al-Madinah Al-Munawarah province, western Saudi Arabia. Int J Infect Dis. 2013;17(5):e334–8.
Haouas N, Amer O, Alshammri FF, Al-Shammari S, Remadi L, Ashankyty I. Cutaneous leishmaniasis in northwestern Saudi Arabia: identification of sand fly fauna and parasites. Parasit Vectors. 2017;10(1):544.
Bern C, Maguire JH, Alvar J. Complexities of assessing the disease burden attributable to leishmaniasis. PLoS Negl Trop Dis. 2008;2:e313.
Alvar J, Bashaye S, Argaw D, Cruz I, Aparicio P, Kassa A, Orfanos G, Parreño F, Babaniyi O, Gudeta N, Cañavate C, Bern C. Kala-azar outbreak in Libo Kemkem, Ethiopia: epidemiologic and parasitologic assessment. Am J Trop Med Hyg. 2007;77(2):275–82.
Ready PD. Leishmaniasis emergence in Europe. Euro Surveill. 2010;15(10):19505.
van Griensven J, Carrillo E, López-Vélez R, Lynen L, Moreno J. Leishmaniasis in immunosuppressed individuals. Clin Microbiol Infect. 2014;20(4):286–99.
Chitimia L, Muñoz-García C, Sánchez-Velasco D, Lizana V, Del Río L, Murcia L, Fisa R, Riera C, Giménez-Font P, Jiménez-Montalbán P, Martínez-Ramírez A, Meseguer-Meseguer JM, García-Bacete I, Sánchez-Isarria MA, Sanchis-Monsonís G, García-Martínez JD, Vicente V, Segovia M, Berriatua E. Cryptic leishmaniosis by Leishmania infantum, a feature of canines only? A study of natural infection in wild rabbits, humans and dogs in southeastern Spain. Vet Parasitol. 2011;181(1):12–6.
Acknowledgements
The authors acknowledged the National Science, Technology and Innovation Plan from King Abdulaziz City for Science and Technology, KSA for providing funding to complete this project. The authors also thank Mr. Casimero A. Victoria for helping in some experimentation.
Funding
This study was funded by the National Science, Technology and Innovation Plan grant from King Abdulaziz City for Science and Technology, KSA (NSTIP/KACST # 11-MED1068–09). NSTIP/KACST participated in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The data used in this study are available and will be provided by the corresponding author on a reasonable request.
Author information
Authors and Affiliations
Contributions
AAAZ, AAAR, MSAD participated in study design, coordination, samples collection and were responsible for obtaining patients written consents. AAA, TS carried out experimentation, statistical analysis and data interpretation. ZR has consulted for data interpretation and manuscript drafting. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was reviewed and approved by the ethics committee of the College of Medicine, Qassim University and the study protocol were approved by NSTIP/KACST from Saudi Arabia. Written informed consent was taken from all studied participants.
Consent for publication
This manuscript does not contain data from any individual person. Consent for publication is “not applicable.”
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Rasheed, Z., Ahmed, A.A., Salem, T. et al. Prevalence of Leishmania species among patients with cutaneous leishmaniasis in Qassim province of Saudi Arabia. BMC Public Health 19, 384 (2019). https://doi.org/10.1186/s12889-019-6710-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12889-019-6710-8