Abstract
Background
The nicotinamide nucleotide transhydrogenase antisense RNA 1 (NNT-AS1) is a long non-coding RNA aberrantly expressed in human malignancies. We aimed to analyze available data to evaluate the correlation between NNT-AS1 expression and cancer prognosis.
Methods
Literature retrieval was performed by systematic searching related databases from inception to April 2, 2020. Studies regarding correlation between NNT-AS1 expression, survival outcomes and clinical characteristics of cancer patients were collected and pooled to calculate the the hazard ratios (HRs) or odds ratios (ORs) with 95% confidence intervals (95% CIs).
Results
Ten studies comprising 699 patients were included, all of which were conducted in China according to literature selection criteria. Overexpression of NNT-AS1 had a significant association with unfavorable overall survival (OS) (HR = 2.08, 95% CI: 1.84–2.36, P < 0.001). Stratified analysis showed that tumor type, sample size, follow-up months, and survival analysis approach did not change the predictive value of NNT-AS1 on OS. Furthermore, elevated NNT-AS1 level had significant association with distant metastasis (DM) (OR = 2.45, 95% CI: 1.39–4.30), lymph node metastasis (LNM) (OR = 3.92, 95% CI: 1.35–11.41), TNM stage (OR = 4.25, 95% CI: 1.71–10.56), and vascular invasion (OR = 3.98, 95% CI: 2.06–7.71), but was not associated with age and gender. The TCGA dataset further consistently showed that the NNT-AS1 expression was associated with poor OS and disease-free survival.
Conclusions
High expression of NNT-AS1 is associated with unfavorable survival outcomes and poor clinicopathologic characteristics. However, large-cohort data and geographical studies are still needed to further validate the prognostic value of NNT-AS1 in cancers.
Similar content being viewed by others
Background
Cancer has become a global health burden and posed a threat to human development over the past decades [1, 2]. Due to cancer, there were 17.2 million incident malignancy cases, 8.9 million deaths, and 213.2 million disability-adjusted life-years worldwide in 2016. Notably, incident cases increased by 28%, of which the largest increase occurred in the least developed countries between 2006 and 2016 [3]. Though tremendous achievements have been made in surgery, chemotherapy, targeted therapy, and the recent immunotherapy in the past years [4, 5], the prognosis of cancer patients still remains poor, which may be ascribed to the lack of effective predictive factors in malignancies. Thus, many investigators have been endeavored to explore novel putative biomarkers for predicting prognosis and therapeutic efficacy in cancer patients [6].
Long non-coding RNAs (lncRNAs) belong to non-coding RNAs whose lengths are longer than 200 bp [7]. They have little or no capability of coding protein [8]. Recent studies have demonstrated that lncRNAs could drive pathophysiologic phenotypes through interaction with other cellular macro-molecules including DNA, RNA and proteins [9]. Aberrant expression or functional abnormalities of LncRNAs have been linked with numerous human diseases, such as aging [10], degenerative disease [11], and cancer [7, 12,13,14]. Recently, a pivotal role of lncRNA in tumor biological characteristics including proliferation, cell cycle arrest, invasion, migration [15], autophagy [16], and drug resistance [17] has also been revealed.
Nicotinamide nucleotide transhydrogenase antisense RNA 1 (NNT-AS1) is a newly identified lncRNA located in the chromosome 5p12 region with three exons, and transcribed in the opposite direction of NNT [18]. Emerging studies have demonstrated that NNT-AS1 could play a crucial role in carcinogenesis, and aberrant expression of NNT-AS1 was significantly associated with survival outcome in various cancers. However, most individual studies evaluating NNT-AS1 expression in cancers remain unconvincing as a result of the limitations in small sample size and possible controversial outcomes. For example, although all eligible studies reported that elevated NNT-AS1 expression was correlated with worse overall survival (OS), study design characteristics such as tumor types, follow-up months, and cut-off values were different among studies. Besides, heterogeneous outcomes regarding clinicopathologic parameters also existed among included studies. For instance, high expression of NNT-AS1 was associated with unfavorable tumor node metastasis (TNM) stage in most studies, but not in study performed by Li Y et al. [19]. Both Ye H et al. [15], and Gu Y et al. [20] reported that upregulated NNT-AS1 indicated positive distant metastasis (DM), but another study conducted by Wang Q et al. [21] showed inconsistent results. Giving the discrepancies and potential underpowered statistical efficacy among those studies, we firstly conducted this comprehensive meta-analysis to elucidate the predictive value of NNT-AS1 in cancer prognosis. This study aimed to provide an objective evaluation on heterogeneity among included studies, and demonstrate the functional mechanisms of NNT-AS1 in carcinogenesis.
Methods
Searching strategy
We searched potential literature MEDLINE, the Cochrane Library, Web of Science, Embase, Scopus, and China National Knowledge Infrastructure (CNKI) database from their inception up to April 2, 2020 to locate articles. In order to strengthen the searching sensitivity, both MeSH terms and free-text words were used. The terms were listed as follows: (“nicotinamide nucleotide transhydrogenase antisense RNA 1” or “NNT-AS1”) AND (“carcinoma” or “sarcoma” or “cancer” or “tumor” or “neoplasm” or “malignancy”) with the limit to human. An additional manual search of citation lists of retrieved literature was performed. Of note, the present study was critically projected, reviewed and reported on the basis of the PRISMA checklist to enhance the credibility of the results [22].
Study selection
For inclusion in the present meta-analysis, the studies should met the following criteria: 1) articles investigating the association between NNT-AS1 expression level and survival outcome in human cancers; 2) patients were categorized into two groups based on the expression of NNT-AS1; 3) patients were diagnosed with cancer by histopathological examination; 4) sufficient original data for extracting or calculating the individual hazard ratios (HRs)/odds ratios (ORs) with its 95% CIs; 5) related clinicopathologic parameters including lymph node metastasis (LNM) and DM were described.
By contrast, studies were excluded according to the following criteria: 1) literature irrelevant to cancer or NNT-AS1; 2) duplicate publications; 3) studies lack of usable clinical data, including animal experiments and those about the structure or functions of NNT-AS1; and 4) letters, editorial, abstracts, case reports or reviews.
Data extraction
All data elements in the enrolled studies were rigorously assessed and extracted by two independent investigators (CT and CHZ), and disagreements were resolved through discussion or consultation from the third investigator (XLR). We extracted the following data from included studies: surname of first author, year of publication, country of origin, tumor type, total number of patients, patients` number in high NNT-AS1 expression group and low NNT-AS1 expression group, clinicopathologic features, detection and survival analysis method, cut-off value, HRs with corresponding 95% CIs regarding to OS, progression-free survival (PFS) or disease-free survival (DFS).
If the data was unavailable, we contacted the corresponding author of original article to request the missing data. When only Kaplan-Meier curves were available in certain studies, the survival rates were indirectly extracted from the graphical plots and calculated HRs with 95% CIs were determined via Engauge Digitizer software (Version 4.1) as previously described [23].
Quality assessment
Two investigators (ZYL and XLR) evaluated the quality of eligible studies independently according to the Newcastle-Ottawa Scale (NOS). Generally, the studies with NOS score ≥ 7 were considered to be of high methodological quality [24] .
Public data and tools
This study is consistent with the publication guidelines provided by The Cancer Genome Atlas (TCGA). TCGA Data portal (https://portal.gdc.cancer.gov) was applied into extracting the clinical data as well as RNAseqV2. Gene Expression Profiling Interactive Analysis (GEPIA) was applied for analysis of the data as described previously [25]. Differential expression analysis was carried out via one-way ANOVA. While the survival analysis, including OS and DFS, were performed by Kaplan–Meier (K-M) and log-rank test, and HRs and p-value were presented as K-M curves.
Statistical methods
All statistical analyses were performed via STATA software (Version 12.0) and Review Manager (RevMan 5.3). Pooled HR with 95% CI was extracted from included studies. The Log HR and standard error (SE) were applied for aggregation of the survival outcomes. Heterogeneity across all studies was determined by I2 statistics and chi-squared test. If I2 > 50% or the chi-squared test shows p < 0.10, which represented significant heterogeneity among the studies, the random-effects model was applied for analysis. In contrary, if apparent between-study heterogeneity was not observed (p > 0.10 and I2 < 50%), the fixed-effects model was adopted.
Sensitivity analysis of NNT-AS1 expression was conducted by sequentially omitting individual study to verify the stability of outcomes in this meta-analysis. The potential publication bias was estimated via Begg’s funnel plot and Egger’s test. If the funnel plot showed asymmetry or Egger’s test showed P < 0.05, the publication bias was considered to be statistically significant.
Results
Characteristics of eligible studies
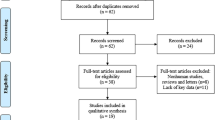
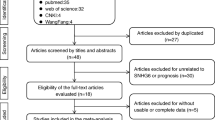
This study was conducted following the PRISMA Checklist, as shown in Table S1. A total of 121 studies were initially identified as potential articles. After removing the duplications, 65 studies were screened through titles and abstracts. Afterwards, 42 articles with irrelevant records were excluded. The remaining 23 full-text articles were further evaluated. Thirteen studies were excluded as a result of irrelevant topics, insufficient data, animal study, and review/ case report. Finally, ten articles compromising 699 patients were included to carry out qualitative and quantitative synthesis. As demonstrated in Fig. 1, the selection procedure was presented by a flow diagram.
The main characteristics of the included studies were demonstrated in Table 1. These articles were published between 2017 and 2019 with a sample size ranging from 42 to 126. In all eligible studies, NNT-AS1 gene expression experiments in this meta-analysis were carried out on primary tumors collected from patients. Generally, the enrolled patients were distributed in two groups (high or low NNT-AS1 expression), considering the levels of NNT-AS1 as measured by qRT-PCR. All of these investigations were carried out in China. Eight divergent types of cancers were analyzed in our meta-analysis, including osteosarcoma, breast cancer, gastric cancers, bladder cancer, cholangiocarcinoma, hepatocellular carcinoma, colorectal cancer and cervical cancer. The follow-up months for survival outcome ranged from 39 to 80 months. Seven studies adopted univariate analysis for the survival analysis method and the other three articles performed multivariate analysis. Furthermore, these studies also investigated other clinicopathologic parameters, such as age, gender, clinical stage, vascular invasion, LNM and DM. As to clinical stage, it should be noted that most studies adopted the TNM classification system, while two studies used the Enneking [15] or the International Federation of Gynecology and Obstetrics (FIGO) staging [30]. All of these eligible studies are of high quality with a NOS score ≥ 7. Details of the NOS scoring were reported in the supplementary file (Table S2).
Association between NNT-AS1 and OS
We used fixed-effects model to analyze the pooled HR and corresponding 95% CI since heterogeneity among these studies was not obvious (I2 = 0.0%, p = 0.932). As presented in Fig. 2a, the pooled result showed that high expression of NNT-AS1 predicted unfavorable OS in cancers (HR = 2.08, 95% CI: 1.84–2.36, P < 0.001).
In addition, stratified analyses were performed to investigate the relevance between NNT-AS1 expression with OS in different subgroups according to tumor type (digestive system or others), sample size (more or less than 60), follow-up months (more or less than 60), and survival analysis method (univariate or multivariate analysis). The results revealed that all stratified analyses recapitulated the predictive potential of NNT-AS1 for OS in malignancies (Fig. 3 and Table 2).
Association between NNT-AS1 and other clinicopathologic parameters
In addition, ORs with corresponding 95% CIs were applied to detect the association between NNT-AS1 and other clinicopathological parameters. The results of these analyses were summarized in Fig. 4 and Table 3. Notably, fixed-effects model was applied in analyzing the association between NNT-AS1 and several clinicopathologic characteristics including age, gender, vascular invasion, and DM, since no obvious heterogeneity was observed. High expression of NNT-AS1 was significantly correlated to vascular invasion (OR = 3.98, 95% CI: 2.06–7.71) and DM (OR = 2.45, 95% CI: 1.39–4.30), but not age and gender.
Forest plots of published articles evaluating the relationship between NNT-AS1 expression and other clinicopathologic features, including a age (≥50/< 50), b age (≥60/< 60), c gender, d clinical stage, e LNM, f DM, and g Vascular invasion. DM: distant metastasis; LNM: lymph node metastasis; NNT-AS1: nicotinamide nucleotide transhydrogenase antisense RNA 1
By contrast, the random-effects model was used to analyze the correlation between NNT-AS1 and clinical characteristics including clinical stage and LNM due to the apparent between-study heterogeneity. Significantly, upregulated expression of NNT-AS1 predicted worse clinical stage (OR = 4.25, 95% CI: 1.71–10.56) and LNM (OR = 3.92, 95% CI: 1.35–11.41).
Sensitivity analysis and publication bias
In order to assess the stability of the aforementioned results, sensitivity analysis was performed. When each eligible study was removed, the result of NNT-AS1 for OS was not obviously changed, indicating the conclusion is reliable (Fig. 2b).
Besides, the publication bias, regarding correlation between expression level of NNT-AS1 and OS, was evaluated via conducting Begg’s funnel plot and Egger’s regression test. The Begg’s funnel plot was symmetry, and Egger’s test showed P = 0.369, suggesting no obvious publication bias was measured (Fig. 2c).
Validation of the result1s in TCGA dataset
Furthermore, the expression levels of NNT-AS1 in related cancers were explored by utilizing the data originated from TCGA. As demonstrated in Fig. 5, NNT-AS1 showed significantly elevated expression in cholangiocarcinoma when compared with control. Besides, NNT-AS1 was aberrantly expressed in sarcoma, stomach adenocarcinoma, liver hepatocellular carcinoma, colon adenocarcinoma, and rectum adenocarcinoma, but the differences were not significant. Moreover, NNT-AS1 expression level was markedly correlated with clinical stage in human cancers. In addition, we merged the expression data and OS/DFS data of carcinomas from TCGA dataset derived from GEPIA, which comprising 4127 patients categorized in high or low expression group, including BLCA (bladder urothelial carcinoma), CESC (cervical squamous cell carcinoma and endocervical adenocarcinoma), BRCA (breast invasive carcinoma), CHOL (cholangiocarcinoma), COAD (colon adenocarcinoma), LIHC (liver hepatocellular carcinoma), LUAD (lung adenocarcinoma), LUSC (lung squamous cell carcinoma), READ (rectum adenocarcinoma), SARC (sarcoma), STAD (stomach adenocarcinoma). These results suggested that the upregulated NNT-AS1 expression predicted worse OS (HR = 1.1, p = 0.018), as well as DFS (HR = 1.1, p = 0.033), confirming that overexpression of NNT-AS1 was significantly correlated to unfavorable survival outcomes in cancer patients.
Validation of NNT-AS1 expression in various cancers in TCGA cohort. a The expression levels of NNT-AS1 in SARC, STAD, LIHC, CHOL, and BRCA. b The expression levels of NNT-AS1 in LUAD, LUSC, COAD, READ, BLCA, and CESC. c Association between NNT-AS1 expression and clinical stage of cancers including SARC, STAD, LIHC, CHOL, COAD, BRCA, LUAD, LUSC, COAD, READ, BLCA, and CESC in TCGA cohort. d OS plot of NNT-AS1 in TCGA cohort (n = 4127). e DFS plot of NNT-AS1 in TCGA cohort (n = 4127). BLCA (bladder urothelial carcinoma); BRCA (breast invasive carcinoma); CESC (cervical squamous cell carcinoma and endocervical adenocarcinoma); CHOL (cholangiocarcinoma); COAD (colon adenocarcinoma); DFS: disease-free survival; LIHC (liver hepatocellular carcinoma); LUAD (lung adenocarcinoma); LUSC (lung squamous cell carcinoma); NNT-AS1: nicotinamide nucleotide transhydrogenase antisense RNA 1; OS: overall survival; READ (rectum adenocarcinoma); SARC (sarcoma); STAD (stomach adenocarcinoma); TCGA: the Cancer Genome Atlas
Discussion
Recently, emerging studies have explored the possible link between expression of lncRNA NNT-AS1 and human tumors. Compared with adjacent noncancerous tissue and normal cell, upregulated NNT-AS1 expression was identified in most cancer tissues or cell lines and therefore indicated poor survival outcome, such as osteosarcoma [31], breast cancer [19], cervical cancer [30], gastric cancer [28], hepatocellular carcinoma [18], colorectal cancer [21], and non-small cell lung cancer (NSCLC) [2]. On the contrary, another study performed by Huang et al, claimed that NNT-AS1 was markedly downregulated in patients with ovarian cancer and ovarian cell lines [32]. However, results from above-mentioned studies should be interpreted with caution because of the limited sample size and discrete outcomes. Therefore, we designed and carried out this meta-analysis to further elucidate the correlation between NNT-AS1 and clinicopathologic outcomes and prognostic values in cancers.
Ten studies with eight cancer types containing 699 patients were pooled together in this study, and the results suggested that promoted NNT-AS1 expression was significantly associated with unfavorable prognosis of OS in patients with cancers. Subgroup stratified analysis further demonstrated that the tumor type, sample size, follow-up months, and survival analysis method did not alter the correlation between TTN-AS1 and OS. Thus, it seems like that differences among study design characteristics have no obvious effect on NNT-AS1 correlation with OS, suggesting that our pooled results were reliable. No publication bias regarding NNT-AS1 expression for OS was observed, indicating the credibility of our results. Furthermore, pooled data from TCGA dataset showed that NNT-AS1 was obviously correlated with OS (HR = 1.1, p = 0.018), which was consistent with our results. Since tumor recurrence is also an imporant prognosis parameter for cancer patients, we next explored the association between NNT-AS1 expression and DFS via TCGA datasets. We showed that NNT-AS1 overexpression was correlated with unfavorable DFS (HR = 1.1, p = 0.033) in the cancer types involved in this meta-analysis (Fig. 5e). Moreover, one study [15] included in our analysis also reported that an elevated expression of TTN-AS1 predicted poor DFS in gastric cancer. However, interpretation regarding DFS should be careful since relevant studies were comparatively limited, and large cohort datasets are still needed for further validation the relationship between NNT-AS1 and tumor recurrence or other parameters. In addition, elevated NNT-AS1 level dramatically predicted worse clinical stage, vascular invasion, LNM, and DM. No significant association between NNT-AS1 and other clinicopathologic parameters including age and gender. Thus, even though NNT-AS1 expression has divergent association with clinicopathological parameters among included studies, our results still provided a reliable correlation via comprehensive meta-analysis. Consistent with our findings, result from the TCGA indicated that NNT-AS1 expression was significantly associated with clinical stage of human cancers. Taken together, our study firstly clarified the relationship between NNT-AS1 and cancer prognosis with a comprehensive evaluation on study heterogeneity and bias. TCGA dataset was explored to validate the role of NNT-AS1 in carcinomas, and the results showed that NNT-AS1 expression may serve as a potential indicator for cancer prognosis.
Previous studies have investigated the underlying mechanisms of NNT-AS1 in carcinogenesis. Overexpression of NNT-AS1showed positive association with poorer OS, advanced tumor stage, LNM, depth of invasion [33], vessel invasion and differentiation in numerous cancers. Functional assays revealed that NNT-AS1 could promote proliferation, weaken cell cycle arrest and alleviate apoptosis by competing with CDK6 for miR-363 binding in hepatocellular carcinoma [18]. High expression of NNT-AS1 facilitates cholangiocarcinoma prognosis via promoting epithelial-mesenchymal transition (EMT) [20]. While knockdown or inhibition of NNT-AS1 could suppress cancer cell colony formation and invasion, arrested the cell cycle and promoted apoptosis both in vitro and in vivo [21]. Additionally, when silencing NNT-AS1 in colorectal cancer, EMT and MAPK/Erk pathway were inhibited [21]. Moreover, other pathways including PI3K/Akt/mTOR and Wnt/β-catenin signaling pathway were also found involved in the tumorigenesis and progression [15, 30]. Besides, NNT-AS1 was capable of serving as a competing endogenous RNA (ceRNA) by sponging miR-485/BCL9 or miR-203 in cholangiocarcinoma [26, 34], miR-1301-3p/PODXL or miR-496/HMGB1 in bladder cancer [3, 27], miR-142-3p/ZEB1 in breast cancer [19], miR-424/E2F1 or miR-363 in gastric cancer [8, 28], miR-22-3p/YAP1 or miR-129-5p in non-small cell lung cancer [33, 35], and miR-320a in osteosarcoma [31], therefore alteration in cancer cell function resulting from NNT-AS1 downregulation may be rescued by miRNA inhibition. Notably, NNT-AS1 also showed a high expression level in drug-resistant NSCLC, which promoted the cisplatin resistance of cancer cells via the MAPK/Slug pathway [36]. All these studies suggested that NNT-AS1 could serve as an oncogenic biomarker in cancer progression. The schematic diagram of various molecules and signaling pathways associated with NNT-AS1 in human cancers were displayed in Fig. 6.
Schematic diagrams of various molecules and signaling pathways associated with NNT-AS1 in human cancers. Aberrant expression of NNT-AS1 was found in various cancers and dysregulation of NNT-AS1 contributed to carcinogenesis through different mechanisms, including promoting cell proliferation, alleviating cell apoptosis, activating invasion and metastasis, promoting EMT and drug-resistance. AKT: protein Kinase B; BCL-2: B-cell lymphoma 2; CDK6: cyclin-dependent kinase 6; E2F1: E2F Transcription Factor 1; EMT: epithelial–mesenchymal transition; ERK: extracellular-signal-regulated kinase; HMGB1: high mobility group box 1; MAPK: mitogen-activated protein kinase; MMP: matrix metalloproteinase; NNT-AS1: nicotinamide nucleotide transhydrogenase antisense RNA 1; PODXL: podocalyxin like; PI3K: phosphoinositide 3-kinase; RUNX2: runt-related transcription factor 2; YAP1: Yes-associated protein 1; ZEB1: zinc finger E-box-binding homeobox 1. Modified from Tamang S’s report [37]
Several deficiencies exist in this meta-analysis and they should be acknowledged. In the first place, our meta-analysis used the summarized data instead of raw data from the specific patients, and most of the HRs and 95% CIs were indirectly calculated by reconstructing survival curves instead of extracted from the original data, which inevitably could cause heterogeneity. Second, the cut-off value for NNT-AS1 expression differed across eligible studies due to the difficulty in reaching a consensus value, thus may introduce possible bias. Third, all enrolled studies were from China, which may cause biased results because of geographical differences. Thus, it should be cautious when applying our conclusions to the population in other regions. Fourth, data regarding NNT-AS1 expression levels with other prognostic outcomes, such as PFS, DFS were limited and thus unable to calculate the pooled value. Fifth, other factors such as different classification system of clinical stage, follow-up time, and analysis methods will also lead to possible bias. Sixth, the regulatory mechanisms of NNT-AS1 in cancer progression still remain largely unexplored. For instance, somatic mutation has been reported to affect lncRNA by regulation of methylation or expression of gene, miRNA, and transcriptional factor [38]. Currently, data regarding the role of somatic mutation on NNT-AS1 is lacking, which may merit future elucidation by using bioinformatics and in vitro assay [38, 39]. Last, in order to further clarify the correlation between NNT-AS1 expression with age or gender, more confounding variables, such as BMI, smoking history, or comorbidities, should be eliminated in order to minimize the possible bias [40]. Therefore, on the basis of the above limitations, well-designed comprehensive studies containing a large sample size, broader regions and countries, and more credible indicators are still warranted to further confirm our results.
Conclusions
In summary, we found that overexpression of NNT-AS1 showed significant association with unfavorable survival outcomes and worse clinicopathological characteristics in kinds of human carcinomas. However, large-cohort data and geographical studies are still needed to conclude a prognostic role of NNT-AS1 as biomarker in cancer.
Availability of data and materials
The data used and analyzed in the study is available from the corresponding author on reasonable request.
Abbreviations
- AKT:
-
Protein Kinase B
- BRCA:
-
Breast invasive carcinoma
- Bcl-2:
-
B-cell lymphoma 2
- CESA:
-
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CDK6:
-
Cyclin-dependent kinase 6
- COAD:
-
Colon adenocarcinoma
- E2F1:
-
E2F Transcription Factor 1
- EMT:
-
Epithelial–mesenchymal transition
- ERK:
-
Extracellular-signal-regulated kinase
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- LIHC:
-
Liver hepatocellular carcinoma
- MAPK:
-
Mitogen-activated protein kinase
- MMP:
-
Matrix metalloproteinase
- mTOR:
-
Mammalian target of rapamycin
- NOS:
-
Newcastle-Ottawa Scale
- NNT-AS1:
-
Nicotinamide nucleotide transhydrogenase antisense RNA1
- OR:
-
Odd ratio
- OS:
-
Overall survival
- PI3K:
-
Phosphoinositide 3-kinase
- RUNX2:
-
Runt-related transcription factor 2
- READ:
-
Rectum adenocarcinoma
- SARC:
-
Sarcoma
- STAD:
-
Stomach adenocarcinoma
- TCGA:
-
The Cancer Genome Atlas
- YAP1:
-
Yes-associated protein 1
- ZEB1:
-
Zinc finger E-box-binding homeobox 1
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Global Burden of Disease Cancer C, Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018;4(11):1553–68.
Sayles LC, Breese MR, Koehne AL, Leung SG, Lee AG, Liu HY, et al. Genome-informed targeted therapy for osteosarcoma. Cancer Discov. 2019;9(1):46–63.
Arab A, Behravan N, Razazn A, Barati N, Mosaffa F, Nicastro J, et al. The viral approach to breast cancer immunotherapy. J Cell Physiol. 2019;234(2):1257–67.
Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8(9):1069–86.
Zhang Y, Lun L, Li H, Wang Q, Lin J, Tian R, et al. The value of lncRNA NEAT1 as a prognostic factor for survival of cancer outcome: a meta-analysis. Sci Rep. 2017;7(1):13080.
Tu C, Ren X, He J, Li S, Qi L, Duan Z, et al. The predictive value of lncRNA MIR31HG expression on clinical outcomes in patients with solid malignant tumors. Cancer Cell Int. 2020;20:115.
Wang X, Ren M, Li Y, et al. Long noncoding RNA NNT-AS1 promotes gastric cancer proliferation and invasion by regulating microRNA-363 expression. J Cell Biochem. 2019;120(4):5704–12.
Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26(2):155–65.
He J, Tu C, Liu Y. Role of lncRNAs in aging and age-related diseases. Aging Med. 2018;1(2):158–75.
Liu Y, Lin L, Zou R, Wen C, Wang Z, Lin F. MSC-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in osteoarthritis. Cell Cycle. 2018;17(21–22):2411–22.
Mehrad-Majd H, Akhtari J, Haerian MS, Ravanshad Y. Clinicopathological and prognostic value of lncRNA PANDAR expression in solid tumors: evidence from a systematic review and meta-analysis. J Cell Physiol. 2019;234(4):4206–16.
Zhang C, Ren X, He J, Wang W, Tu C, Li Z. The prognostic value of long noncoding RNA SNHG16 on clinical outcomes in human cancers: a systematic review and meta-analysis. Cancer Cell Int. 2019;19:261.
Tu C, Ren X, He J, Zhang C, Chen R, Wang W, et al. The value of LncRNA BCAR4 as a prognostic biomarker on clinical outcomes in human cancers. J Cancer. 2019;10(24):5992–6002.
Ye H, Lin J, Yao X, Li Y, Lin X, Lu H. Overexpression of long non-coding RNA NNT-AS1 correlates with tumor progression and poor prognosis in osteosarcoma. Cell Physiol Biochem. 2018;45(5):1904–14.
Xiong H, Ni Z, He J, Jiang S, Li X, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36(25):3528–40.
Corra F, Agnoletto C, Minotti L, Baldassari F, Volinia S. The network of non-coding RNAs in cancer drug resistance. Front Oncol. 2018;8:327.
Lu YB, Jiang Q, Yang MY, Zhou JX, Zhang Q. Long noncoding RNA NNT-AS1 promotes hepatocellular carcinoma progression and metastasis through miR-363/CDK6 axis. Oncotarget. 2017;8(51):88804–14.
Li Y, Lv M, Song Z, Lou Z, Wang R, Zhuang M. Long non-coding RNA NNT-AS1 affects progression of breast cancer through miR-142-3p/ZEB1 axis. Biomed Pharmacother. 2018;103:939–46.
Gu Y, Li C, Xiao L, Li J, Pei H, Xu D, et al. High expression of long non-coding RNA NNT-AS1 facilitates progression of cholangiocarcinoma through promoting epithelial-mesenchymal transition. Am J Transl Res. 2019;11(9):5438–56.
Wang Q, Yang L, Hu X, Jiang Y, Hu Y, Liu Z, et al. Upregulated NNT-AS1, a long noncoding RNA, contributes to proliferation and migration of colorectal cancer cells in vitro and in vivo. Oncotarget. 2017;8(2):3441–53.
Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4(1):1.
Xu L, Yan N, Li Z, Luo L, Wu X, Liu Q, et al. A comparison of fulvestrant plus a targeted agent with fulvestrant alone in hormone receptor-positive advanced breast cancer that progressed on prior endocrine therapy: a meta-analysis. OncoTargets Ther. 2018;11:8389–98.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102.
Huang L, Jiang X, Kang P, Wang Z, Leng K, Ji D, et al. Long non-coding RNA NNT-AS1 functions as an oncogenic gene through modulating miR-485/BCL9 in cholangiocarcinoma. Cancer Manag Res. 2019;11:7739–49.
Wu D, Zhang T, Wang J, Zhou J, Pan H, Qu P. Long noncoding RNA NNT-AS1 enhances the malignant phenotype of bladder cancer by acting as a competing endogenous RNA on microRNA-496 thereby increasing HMGB1 expression. Aging (Albany NY). 2019;11(24):12624–40.
Chen B, Zhao Q, Guan L, Lv H, Bie L, Huang J, et al. Long non-coding RNA NNT-AS1 sponges miR-424/E2F1 to promote the tumorigenesis and cell cycle progression of gastric cancer. J Cell Mol Med. 2018;22(10):4751–9.
Gu Youhua XF, Xueping Y, Jian Z, Yueju W. Expression and clinical significance of LncRNA NNT-AS1 in gastric carcinoma. Shandong Med J. 2018;58(41):18–21.
Hua F, Liu S, Zhu L, Ma N, Jiang S, Yang J. Highly expressed long non-coding RNA NNT-AS1 promotes cell proliferation and invasion through Wnt/beta-catenin signaling pathway in cervical cancer. Biomed Pharmacother. 2017;92:1128–34.
Li C, Zhang S, Qiu T, Wang Y, Ricketts DM, Qi C. Upregulation of long non-coding RNA NNT-AS1 promotes osteosarcoma progression by inhibiting the tumor suppressive miR-320a. Cancer Biol Ther. 2019;20(4):413–22.
Huang Y, Shi J, Xu Y. Long non-coding RNA NNT-AS1 contributes to cell proliferation, metastasis and apoptosis in human ovarian cancer. Oncol Lett. 2018;15(6):9264–70.
He W, Zhang Y, Xia S. LncRNA NNT-AS1 promotes non-small cell lung cancer progression through regulating miR-22-3p/YAP1 axis. Thorac Cancer. 2020;11(3):549–60.
Gu Y, Zhu Z, Pei H, Xu D, Jiang Y, Zhang L, et al. Long non-coding RNA NNT-AS1 promotes cholangiocarcinoma cells proliferation and epithelial-to-mesenchymal transition through down-regulating miR-203. Aging (Albany NY). 2020;12(3):2333–46.
Shen Q, Jiang Y. LncRNA NNT-AS1 promotes the proliferation, and invasion of lung cancer cells via regulating miR-129-5p expression. Biomed Pharmacother. 2018;105:176–81.
Cai YDZ, Wang JY. LncRNA NNT-AS1 is a major mediator of cisplatin chemoresistance in non-small cell lung cancer through MAPK/slug pathway. Eur Rev Med Pharmacol Sci. 2018;22(15):9.
Tamang S, Acharya V, Roy D, Sharma R, Aryaa A, Sharma U, et al. SNHG12: an LncRNA as a potential therapeutic target and biomarker for human Cancer. Front Oncol. 2019;9:901.
Gao Y, Li X, Zhi H, Zhang Y, Wang P, Wang Y, et al. Comprehensive characterization of somatic mutations impacting lncRNA expression for Pan-cancer. Mol Ther Nucleic Acids. 2019;18:66–79.
Minotti L, Agnoletto C, Baldassari F, Corra F, Volinia S. SNPs and somatic mutation on long non-coding RNA: new frontier in the cancer studies? High Throughput. 2018;7(4):34.
Care A, Bellenghi M, Matarrese P, Gabriele L, Salvioli S, Malorni W. Sex disparity in cancer: roles of microRNAs and related functional players. Cell Death Differ. 2018;25(3):477–85.
Acknowledgements
Not Applicable.
Funding
This work was supported by the National Natural Science Foundation of China 81902745); Natural Science Foundation of Hunan Province (2018JJ3716); and Central South University Innovative Program for Undergraduates (No. 20190034020002). The funding parties had no role in the study design, data collection and analysis, and interpretation of data, or in the manuscript writing.
Author information
Authors and Affiliations
Contributions
Study design: CT; Data collection and analysis: CT and CHZ; Quality assessment: XLR and ZYL; Manuscript preparation and revision: CT and CHZ; Supervision of the project: CT; Final approval of the manuscript: All authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Chao Tu is a member of the editorial board of BMC Cancer. The authors declare that they approve this article and have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
PRISMA Checklist.
Additional file 2: Table S2.
Study quality and bias in the retrospective cohort studies judged by the Newcastle-Ottawa Scale (NOS) checklist.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, C., Ren, X., Liu, Z. et al. Upregulated expression of LncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1 is correlated with unfavorable clinical outcomes in cancers. BMC Cancer 20, 879 (2020). https://doi.org/10.1186/s12885-020-07348-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-020-07348-5