Abstract
Background
Prognostication of patients with colorectal cancer (CRC) currently relies on tumor-node-metastasis (TNM) staging but clinical outcomes of patients of the same histoclinical stage are heterogeneous. It is therefore imperative to devise novel molecular tests to stratify CRC patients. Our previous work demonstrated that eukaryotic elongation factor-2 kinase (EEF2K) is a tumor suppressor in CRC. Herein, we investigated EEF2K expression in CRC and determined its relationship with clinicopathological parameters.
Methods
Quantitative RT-PCR and Westerns blots were used to examine EEF2K expression in primary tumor and the adjacent non-tumor tissues of CRC patients (n = 20). Kaplan-Meier curves and Cox regression analysis were used to assess the association between clinical outcomes of CRC patients and EEF2K protein expression determined by immunohistochemistry on tissue microarray (n = 151).
Results
EEF2K was significantly downregulated at both mRNA and protein levels in tumors of CRC patients. Univariate Cox regression analysis revealed that CRC patients with high tumor grade, advanced TNM staging and low EEF2K expression were associated with worse overall survival. Multivariate analysis further demonstrated that low EEF2K expression was an independent factor for predicting poorer overall survival in CRC patients (p = 0.014; Hazard ratio = 2.951; 95% confidence interval: 1.240–7.024). The 5-year survival rate was 82.8% in the EEF2K-high-expression group versus 63.9% in the EEF2K-low-expression group (p = 0.0118). The association of overall survival with EEF2K expression in CRC patients was verified in The Cancer Genome Atlas (TCGA) cohort.
Conclusions
EEF2K is downregulated in CRC and its expression can be employed as a prognostic marker for CRC patients independent of TNM staging.
Similar content being viewed by others
Background
Colorectal cancer (CRC) is the second most common malignant disease and the fourth leading cause of cancer-related death worldwide, accounting for more than 650,000 deaths annually [1]. In particular, CRC incidence in many Asian countries has increased 2- to 4-fold over the last two decades [2]. Despite the recent advancements in CRC treatment, up to 50% patients whom underwent tumor resection experience cancer recurrence, among which another half subsequently developed virtually incurable metastasis. The prognostication of CRC patients remains a clinical challenge with tumor-node-metastasis (TNM) staging as the most commonly used prognostic tool in the clinical setting. However, the clinical value of this system in guiding suitable therapy has recently been questioned. For instance, adjuvant therapy is recommended for all stage III patients with CRC but it remains controversial for stage II patients as its toxicities may outweigh benefits. It is therefore pivotal to identify early-stage CRC patients with predicted gloomy outcomes pending for more aggressive treatment. In this respect, the utility of molecular predictive and prognostic markers has helped forecast clinical outcome and treatment responsiveness for deciding better interventions for CRC patients. For example, KRAS mutation has been established as a negative predictor for response to epidermal growth factor receptor-targeted therapy in patients with metastatic CRC [3]. Also, programmed death (PD)-ligand 1 (L1) expression on tumor cells predicts efficacy of PD-L1 and PD-1 inhibition-based therapies in different types of cancer [4]. Thus, the development of novel prognostic marker for clinical outcome prediction in CRC is highly warranted.
Eukaryotic elongation factor-2 kinase (EEF2K) is a calcium/calmodulin-dependent protein kinase that plays a role in regulating protein synthesis [5]. EEF2K phosphorylates its downstream target eukaryotic elongation factor-2 (EEF2), a GTPase that promotes the translocation of the nascent protein chain from A site to P site on ribosome in elongation phase during mRNA translation [6]. The phosphorylation of EEF2 at threonine 56 by EEF2K inhibits the interaction of EEF2 with ribosome and thereby inhibiting protein elongation. In our recent study, a tumor-suppressive role of EEF2K was observed in CRC, where silencing of EEF2K induced a pro-survival autophagic response through the AMPK-ULK pathway and promoted CRC growth through increasing cell size, viability and clonogenicity. In contrast, overexpressing EEF2K reduced colon cancer cell viability and potentiated the anti-tumor efficacy of the chemotherapeutic drug oxaliplatin [7]. Consistently, De Gassart and co-workers reported that nelfinavir exerted its anti-tumor effect in colon cancer cells in an EEF2K-dependent manner [8]. An in vivo study by Faller and his colleagues also revealed that EEF2K activation led to growth arrest in APC-deficient colorectal adenomas [9]. All these pieces of evidence support EEF2K as a tumor-suppressor gene in CRC. However, the clinical significance of EEF2K downregulation in CRC remains to be established.
In the current study, we investigated the expression of EEF2K in CRC and delineated its relationship with clinicopathological parameters, including survival, in patients with CRC.
Methods
Clinical samples
Twenty pairs of primary tumor tissues and matched adjacent non-tumor tissues were collected during operation from CRC patients who were admitted to the Prince of Wales Hospital, Shatin, Hong Kong. All specimens were immediately frozen in liquid nitrogen and stored at -80 °C until use. For tissue microarrays, formalin-fixed, paraffin-embedded archived CRC tissues were used. Use of these tissues had been approved by the Joint Chinese University of Hong Kong—New Territories East Cluster Clinical Research Ethics Committee. Informed written consents from patients were obtained.
The Cancer Genome Atlas (TCGA) cohort
Expression data and clinical information were collected from the TCGA open access data directory. Reads mapped to EEF2K (level 3 data) were used to quantify EEF2K mRNA expression levels and normalized by transcripts per million mapped reads (TPM).
RNA extraction, reverse transcription and real-time PCR
Total RNA was extracted from aforementioned matched samples using TRIzol reagent (Life Technologies) following manufacturer’s protocol. First-strand complementary DNA was synthesized from the extracted RNA using PrimeScript™ RT Reagent kit (TaKaRa) with a protocol suggested by the manufacturer. EEF2K levels were quantified using Power SYBR® Green PCR master mix (Applied Biosciences) on ABI Quantstudio™ 7 Flex Real Time PCR System (Thermo-cycling condition: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min). Relative expression was calculated using 2-∆∆Ct method using β-actin for normalization.
Protein extraction and Western blots
Total protein was extracted by homogenizing the tissues in radioimmunoprecipitation assay buffer (150 mM sodium chloride, 50 mM Tris, 1% Triton-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate, pH 7.6) containing PhosSTOP™ phosphatase inhibitors (Roche) and cocktail protease inhibitors (Sigma). After centrifugation at 12,000⨯g for 20 min, the supernatant was mixed and heated with Laemmli buffer. Proteins were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane (Pall Corporation), which was then incubated with antibodies against EEF2K (Cell Signaling Technology) or β-actin (Santa Cruz Biotechnology) at 4 °C overnight. The membranes were then washed and incubated with horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology) and the bands were visualized with enhanced chemiluminescent reagent (Advansta).
Immunohistochemistry for EEF2K
Tissue microarrays comprising formalin-fixed paraffin-embedded tumor tissue sections from 160 cases of CRC were subjected to immunohistochemistry for determining EEF2K protein levels using Histostain® Plus LAB-SA Detection System (ThermoFisher) with a slightly modified protocol. Briefly, tissue sections were deparaffinized and rehydrated, and incubated in 3% hydrogen peroxide for 10 min to block endogenous peroxidase activity. Antigens were retrieved by microwave in sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) for 5 min. After cooling to room temperature, sections were incubated in 10% goat serum (Dako) for 2 h and then with antibody against EEF2K (Novartis) at 4 °C overnight, and substantially washed with phosphate-buffered saline with 0.025% Tween 20. Then, sections were incubated with biotinylated secondary antibodies, followed by streptavidin-conjugated horseradish peroxidase. After washing, signals were developed using 3, 3′-diaminobenzidine as the chromogen and counterstained with hematoxylin. Expression levels of EEF2K on each specimen were evaluated and scored according to the percentage of epithelial cells possessing positive signals and the intensity of the positive signals. Percentage of positive signal was graded as: 1 (5–10%); 2 (11–20%); 3 (> 20%). Intensity of signal was graded as: 0 (none); 1 (weak); 2 (moderate); 3 (strong). EEF2K score was calculated as the product of the two grades. Sections with score ≥ 2 were classified as EEF2K-positive and < 2 as EEF2K-negative.
Statistical analysis
Chi-square test was used to determine the correlation between grouped EEF2K expression with categorical clinicopathological parameters. Student’s t test and one-way analysis of variance was used to compare means of continuous data. Kaplan-Meier analysis and log-rank test were used to evaluate the correlation of EEF2K expression with patients’ survival. Univariate and multivariate Cox regression analyses were used to assess the prognostic value of EEF2K expression and other clinicopathological parameters in predicting survival outcome. The proportional hazard assumption of the Cox analysis was verified by assessing the scaled Schoenfeld residuals and time-dependent covariates for each predictor. Survival-related statistical analyses were conducted using SPSS 22.0 software while other statistical tests were conducted with GraphPad Prism 6. Results were considered significant when two-sided p value < 0.05. Experiments were conducted for at least 3 times.
Results
EEF2K was downregulated in CRC at mRNA and protein levels
To unravel the potential dysregulation of EEF2K in CRC tissues, the relative mRNA and protein expression levels of EEF2K were assessed by real-time PCR and Western blots, respectively. Eighty-five percent of the cases (17 out of 20) showed a more than two-fold downregulation of EEF2K mRNA in cancerous tissues as compared to the adjacent non-tumor tissues (Fig. 1a). In general, the downregulation of EEF2K in tumor tissues was significant (Fig. 1b). At protein level, consistent with the mRNA analysis, EEF2K was significantly reduced in 75% of the cases (15 out of 20) in primary tumor tissue relative to the paired non-tumor tissues (Fig. 1c). The downregulation of EEF2K in CRC tissues was not associated with patients’ age, gender, tumor location, tumor grade, tumor stage or nodal stage (Table 1).
Downregulation of EEF2K in human CRC. a Individual and b overall EEF2K mRNA expression in 20 pairs of CRC tissue and paired non-tumor tissue evaluated by real-time PCR analysis. Negative value denotes downregulation in tumor tissue over normal counterparts. c Western blots of EEF2K protein in 20 pairs of matched adjacent non-tumor (N) and tumor (T) tissues. Quantification was performed using ImageJ software and expressed as ratio of T over N for each pair of tissue. ***, p < 0.001 against adjacent non-tumor tissue by two-sided paired t-test
Immunohistochemical staining assessment and correlation analysis of EEF2K expression with patients’ clinicopathological factors
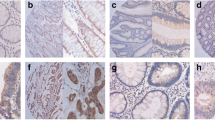
Tissue microarrays comprising 160 CRC tissues were subjected to immunohistochemistry for detecting EEF2K protein expression. In addition to tissues lost during staining and samples with incomplete information, patients who survived less than 3 months after surgery were filtered out to exclude death caused by surgical complications. Finally, 151 cases were enrolled for the current analysis, of which 19.2% cases (29 out of 151) were classified as EEF2K-positive and 80.8% cases (122 out of 151) as EEF2K-negative based on percentage and intensity of positive staining (Fig. 2). EEF2K expression was not correlated with patients’ age, gender, tumor location, tumor grade, TNM staging or microsatellite instability (MSI) status (Table 2).
Low EEF2K expression was correlated with worse clinical outcome
To evaluate the clinical relevance of EEF2K expression in CRC, we examined the correlation of EEF2K expression with clinical outcomes of the patients in our cohort. Kaplan-Meier analysis showed that patients in the EEF2K-negative group exhibited significantly worse overall survival than those in the EEF2K-positive group (Fig. 3). We then assessed the prognostic potential of EEF2K expression in predicting overall survival with Cox regression analysis. Univariate analysis of our patient cohort indicated that high tumor grade, advanced TNM staging and low EEF2K expression were predictors for shortened overall survival. In multivariate analysis, low EEF2K expression was shown to be an independent predictor for poorer overall survival (Hazard ratio = 2.951, with reference to EEF2K positive group; 95% confidence interval: 1.240–7.024; p = 0.014) after controlling tumor grade, TNM staging together with age and gender of the patients for potential confounding effects (Table 3).
Validating the expression and prognostic significance of EEF2K in TCGA cohort
The expression of EEF2K and its association with overall survival of CRC patients were further studied using The Cancer Genome Atlas (TCGA) cohort. Data of RNA sequencing, together with the clinical features and survival information of CRC patients, were obtained from the TCGA Data Portal. Our analysis demonstrated that EEF2K expression was significantly downregulated in CRC in both unpaired and paired comparison between normal and tumor tissues (Fig. 4). The downregulation of EEF2K was common but not associated with patients’ clinical characteristics (age, gender, tumor location, TNM staging), CRC subtypes (sporadic vs familial) or mutational status (MSI status and mutation of KRAS, BRAF, APC and TP53) (Fig. 5 and Additional file 1). The clinical relevance of EEF2K expression in CRC patients was then evaluated. Patients with the highest 20% EEF2K mRNA expression (high expression group) and the lowest 20% (low expression group) among the cohort were selected and their survival outcomes were compared (Additional file 2). Our analysis indicated that, after stratifying at stage IV CRC (AJCC), patients in the low expression group had significantly worse overall survival than those in the high expression group (20 cases; Fig. 6).
Association of EEF2K expression with patients’ clinicopathological factors and mutational status in TCGA cohort. EEF2K expression was obtained from TCGA RNA sequencing data and expressed as transcripts per million mapped reads (TPM). P values were obtained by student’s t test and one-way analysis of variance when comparing factors of two groups and more than two groups respectively. The whisker spans the 95% confidence interval
Discussion
EEF2K, together with other enzymes involved in protein synthesis, were traditionally known as cellular housekeepers that play a vital role in cell viability [10]. In our study, we further extended the role of EEF2K as a prognostic biomarker for CRC in which low EEF2K expression foreshadowed worse overall survival independent of other clinicopathological parameters, including age, gender and TNM staging of the patients. This observation is consistent with our previous finding that EEF2K functioned as a tumor-suppressor gene in CRC by inhibiting autophagic survival and synergized with oxaliplatin (a commonly used chemotherapeutic drug in CRC management) to induce colon cancer cell apoptosis [7]. Our findings were further validated in TCGA cohort, in which EEF2K was also downregulated in CRC without association with patients’ clinical features, CRC subtypes or mutational status as in our cohort. However, its clinical relevance with survival outcome of CRC patients was only noted in patients with stage IV CRC. It is aware that the methodologies in assessing EEF2K expression level, and also the ethnicity of patients and underlying genetic background were different and may account for the discrepancy. Also, it should be noted that multiple comparisons were not adjusted for this analysis. Additional dedicated studies are needed for further confirmation. Despite all these, our findings indicated that EEF2K expression, alongside other molecular prognostic markers such as microsatellite instability [11], BRAF mutation status [12], gene expression profiling [13] and multi-gene mutation signature [14], might supplement TNM staging in the future for more accurate prognostication and patient stratification.
The dysregulation of EEF2K and its clinical relevance have been documented in other cancer types. It is noteworthy that, contrary to its tumor-suppressive role in CRC, EEF2K was frequently found to function as an oncogene in other cancer types. Ashour and colleagues reported an overexpression of EEF2K in pancreatic cancer [15]. Meric-Bernstam and colleagues found that EEF2K was overexpressed in breast cancer and, importantly, the expression of EEF2K was positively correlated with poor prognosis in breast cancer patients [16]. A similar finding was also reported in brain cancer by Leprivier and his colleagues [17]. The tissue-specific functions of EEF2K may provide a plausible explanation for such discrepancy. While EEF2K overexpression has been shown to promote cancer cell survival in face of nutrient starvation by inhibiting protein synthesis resulted in limitation of cellular energy exhaustion [18], mTOR complex 1-mediated inhibition of EEF2K was found to be essential for the proliferation of APC-deficient but not wild-type enterocytes [9]. This distinctive synthetic lethality circuit created by APC inactivation in CRC strongly supports the intestine-specific tumor-suppressive effect of EEF2K and its association with better prognosis in CRC patients. This finding also suggests that EEF2K may function as a tumor suppressor in other cancer types, such as endometrial [19] and gastric cancers [20], in which APC inactivation plays a significant oncogenic role. The prognostic role of EEF2K expression in these cancer types also warrants further investigation to support the notion.
Conclusions
Taken together, we found that EEF2K downregulation is independently associated with worse overall survival in CRC patients. To the best of our knowledge, it is the first time demonstrating the clinical relevance of EEF2K expression in CRC patients. The use of EEF2K as a prognostic marker could be promising in identifying high-risk CRC patients to improve their survival with more aggressive treatment.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CRC:
-
Colorectal cancer
- EEF2:
-
Eukaryotic elongation factor-2
- EEF2K:
-
Eukaryotic elongation factor-2 kinase
- MSI:
-
Microsatellite instability
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- TNM:
-
Tumor-node-metastasis
References
Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray, F. GLOBOCAN 2012 v1.0, Cancer incidence and mortality worldwide: IARC CancerBase no. 11 [internet]. Lyon, France: International Agency for Research on Cancer; 2013. Available from: http://gco.iarc.fr. Accessed 25 Dec 2016.
Sung JJ, Lau JY, Goh KL, Leung WK. Asia Pacific Working Group on Colorectal Cancer. Increasing incidence of colorectal cancer in Asia: implications for screening. Lancet Oncol. 2005;6(11):871–6.
Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101(19):1308–24.
Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17(12):e542–51.
Nairn AC, Bhagat B, Palfrey HC. Identification of calmodulin-dependent protein kinase III and its major MR 100,000 substrate in mammalian tissues. Proc Natl Acad Sci U S A. 1985;82(23):7939–43.
Carlberg U, Nilsson A, Nygård O. Functional properties of phosphorylated elongation factor 2. Eur J Biochem. 1990;191(3):639–45.
Xie CM, Liu XY, Sham KWY, Lai JMY, Cheng CHK. Silencing of EEF2K (eukaryotic elongation factor-2 kinase) reveals AMPK-ULK1-dependent autophagy in colon cancer cells. Autophagy. 2014;10(9):1495–508.
De Gassart A, Demaria O, Panes R, Zaffalon L, Ryazanov AG, Gilliet M, Martinon F. Pharmacological eEF2K activation promotes cell death and inhibits cancer progression. EMBO Rep. 2016;17(10):1471–84.
Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, Dudek KM, Casey HA, Scopelliti A, Cordero JB, Vidal M, Pende M, Ryazanov AG, Sonenberg N, Meyuhas O, Hall MN, Bushell M, Willis AE, Sansom OJ. mTORC1-mediated translational elongation limits intestinal tumor initiation and growth. Nature. 2015;517(7535):497–500.
Thornton S, Anand N, Purcell D, Lee J. Not just for housekeeping: protein initiation and elongation factors in cell growth and tumorigenesis. J Mol Med (Berl). 2003;81(9):536–48.
Merok MA, Ahlquist T, Røyrvik EC, Tufteland KF, Hektoen M, Sjo OH, Mala T, Svindland A, Lothe RA, Nesbakken A. Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: results from a large, consecutive Norwegian series. Ann Oncol. 2013;24(5):1274–82.
Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, Slattery ML. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65(14):6063–9.
Nannini M, Pantaleo MA, Maleddu A, Astolfi A, Formica S, Biasco G. Gene expression profiling in colorectal cancer using microarray technologies: results and perspectives. Cancer Treat Rev. 2009;35(3):201–9.
Yu J, Wu WK, Li X, He J, Li XX, Ng SS, Yu C, Gao Z, Yang J, Li M, Wang Q, Liang Q, Pan Y, Tong JH, To KF, Wong N, Zhang N, Chen J, Lu Y, Lai PB, Chan FK, Li Y, Kung HF, Yang H, Wang J, Sung JJ. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut. 2015;64(4):636–45.
Ashour AA, Abdel-Aziz AA, Mansour AM, Alpay SN, Huo L, Ozpolat B. Targeting elongation factor-2 kinase (eEF-2K) induces apoptosis in human pancreatic cancer cells. Apoptosis. 2014;19(1):241–58.
Meric-Bernstam F, Chen H, Akcakanat A, Do KA, Lluch A, Hennessy BT, Hortobagyi GN, Mills GB, Gonzalez-Angulo A. Aberrations in translational regulation are associated with poor prognosis in hormone receptor-positive breast cancer. Breast Cancer Res. 2012;14(5):R138.
Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, Agnihotri S, El-Naggar A, Yu B, Somasekharan SP, Faubert B, Bridon G, Tognon CE, Mathers J, Thomas R, Li A, Barokas A, Kwok B, Bowden M, Smith S, Wu X, Korshunov A, Hielscher T, Northcott PA, Galpin JD, Ahern CA, Wang Y, McCabe MG, Collins VP, Jones RG, Pollak M, Delattre O, Gleave ME, Jan E, Pfister SM, Proud CG, Derry WB, Taylor MD, Sorensen PH. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153(5):1064–79.
Moore CE, Wang X, Xie J, Pickford J, Barron J, Regufe da Mota S, Versele M, Proud CG. Elongation factor 2 kinase promotes cell survival by inhibiting protein synthesis without inducing autophagy. Cell Signal. 2016;28(4):284–93.
Moreno-Bueno G, Hardisson D, Sánchez C, Sarrió D, Cassia R, García-Rostán G, Prat J, Guo M, Herman JG, Matías-Guiu X, Esteller M, Palacios J. Abnormalities of the APC/beta-catenin pathway in endometrial cancer. Oncogene. 2002;21(52):7981–90.
Nakatsuru S, Yanagisawa A, Ichii S, Tahara E, Kato Y, Nakamura Y, Horii A. Somatic mutation of the APC gene in gastric cancer: frequent mutations in very well differentiated adenocarcinoma and signet-ring cell carcinoma. Hum Mol Genet. 1992;1(8):559–63.
Acknowledgements
Not applicable.
Funding
This work was supported by the Chinese University of Hong Kong. The authors also thank the University for the provision of postgraduate studentships. The Chinese University of Hong Kong was not involved in the design of the study, collection, analysis or interpretation of data, writing of this manuscript or decision to submit for publication.
Author information
Authors and Affiliations
Contributions
THN drafted the manuscript. THN, LZ, JHMT and WYZL conducted the experiments. THN, KWYS, CMX and MTVC analyzed the data. KFT and SSMN collected clinical specimens and information. WKKW and CHKC designed and supervised the study and revised the manuscript. All authors have read and approved this manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Informed written consents from patients were obtained. Use of tissues had been approved by the Joint Chinese University of Hong Kong—New Territories East Cluster Clinical Research Ethics Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Association of EEF2K expression with BRAF, APC and TP53 gene mutation in TCGA cohort. EEF2K expression was obtained from TCGA RNA sequencing data. P values were obtained by student’s t test. The whisker spans the 95% confidence interval. (TIF 220 kb)
Additional file 2:
Significance levels and different grouping cut-offs of EEF2K expression in survival analysis. Analysis was performed from data in TCGA cohort. Patients were grouped into high and low EEF2K expression groups by different cut-off values of EEF2K expression based on percentiles. nth percentile indicates that patients fall into the lowest n% and highest n% in EEF2K expression were selected and grouped into low expression and high expression group respectively. Their overall survivals were compared and significance levels (or p values) were obtained by log-rank tests accordingly. **, p < 0.01. (TIF 90 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ng, T.H., Sham, K.W.Y., Xie, C.M. et al. Eukaryotic elongation factor-2 kinase expression is an independent prognostic factor in colorectal cancer. BMC Cancer 19, 649 (2019). https://doi.org/10.1186/s12885-019-5873-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-019-5873-0