Abstract
Background
Cetuximab, an anti-EGFR monoclonal antibody, is used in combination with chemotherapy in clinic to enhance the outcome in metastatic colorectal cancer (mCRC) patients with only ~ 20% response rate. To date only activating mutations in KRAS and NRAS have been identified as poor prognosis biomarkers in cetuximab-based treatment, which makes an urgent need for identification of novel prognosis biomarkers to precisely predict patients’ response in order to maximize the benefit.
Methods
In this study, we analysed the mutation profiles of 33 Chinese mCRC patients using comprehensive next-generation sequencing (NGS) targeting 416 cancer-relevant genes before cetuximab treatment. Upon receiving cetuximab-based therapy, patients were evaluated for drug response, and the progression-free survival (PFS) was monitored. The association of specific genetic alterations and cetuximab efficacy was analyzed.
Results
Patients carrying SMAD4 mutations (SMAD4mut, n = 8) or NF1 mutations (NF1mut, n = 4) had significantly shorter PFS comparing to those carrying wildtype SMAD4 (SMAD4wt, n = 25) (P = 0.0081) or wildtype NF1 (NF1wt, n = 29) (P = 0.0028), respectively. None of the SMAD4mut or NF1mut patients showed response to cetuximab when assessed at 12-week post-treatment. Interestingly, two patients carrying both SMAD4mut and NF1mut showed the shortest PFS among all the patients.
Conclusions
Our results demonstrated that SMAD4 and NF1 mutations can serve as potential biomarkers for poor prognosis to cetuximab-based therapy in Chinese mCRC patients.
Similar content being viewed by others
Background
Colorectal cancer (CRC) represents a major public health issue due to its high incidence and mortality rate. In China, it is the fifth most common cancer in 2015 and causes ~ 191,000 deaths annually [1]. Comparing to other malignant tumors, CRC has a higher intra-tumoral heterogeneity and harbours higher tumor mutation burden [2, 3]. Common driver genes for CRC include TP53, APC, KRAS, NRAS, PIK3CA, and SMAD4, etc. [4, 5].
Current treatment of unresectable metastatic CRC (mCRC) in advanced stage mainly relies on fluoropyrimidine-based chemotherapies alone or in combination with Bevacizumab according to the National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology (NCCN Guidelines) in Colon Cancer [6, 7]. The efficacy of chemotherapy could be further increased via the addition of cetuximab or panitumumab, which are humanized monoclonal antibodies that target the extracellular ligand binding domain of the epidermal growth factor receptor (EGFR) [8, 9]. However, only ~ 20% of patients benefit from the cetuximab-based therapy [10]. To date, prognostic biomarkers for cetuximab treatment is limited to the activating mutations of KRAS and NRAS, which have been approved for poor response to cetuximab [11,12,13]. Other potential biomarkers, including mutations in BRAF (V600E), PIK3CA, SMAD4, PTEN, etc., have also been reported, albeit requiring further validation [14,15,16,17]. However, all the aforementioned biomarkers could not adequately explain the poor response rate, and thus limits patients to get maximal benefit from cetuximab-based therapy.
The aim of this study was to seek better prognosis biomarkers for cetuximab-based therapy via a comprehensive analysis of the mCRC patients’ mutation profiles using next-generation sequencing (NGS) targeting 416 cancer-relevant genes. In combination with their response to cetuximab treatment, our data show that patients carrying at least one of SMAD4 or NF1 mutations had a higher possibility of a poor response to EGFR blockade with a shorter progression-free survival (PFS), suggesting that SMAD4 and NF1 mutations may play an important role in tumor progression and might function as biomarkers for poor prognosis to cetuximab-based therapy in mCRC patients.
Methods
Patients
This study was approved by the ethic committee of the First Affiliated Hospital with Nanjing Medical University. From 2009 to Nov 2015, tumor specimens or plasma samples were collected at the time of diagnosis from 33 Chinese patients with mCRC with written informed consents. All patients received at least one course of cetuximab treatment in combined with FOLFIRI, FOLFOX or XELOX until progressive disease (PD) was observed. Disease progression was evaluated every 6 weeks during treatment according to the Response Evaluation Criteria In Solid Tumors (RECIST) version 1.1 [18]. PFS was calculated from the first day of the administration of cetuximab-based therapy until PD. According to the Guidance for Industry Clinical Trial Endpoints for the Approval of Cancer Drugs and Biologics, partial response (PR) and complete response (CR) are considered as response to the treatment applied [19].
Clinical sample collection and DNA extraction
Specimen collection and preparation were performed following the standard protocols approved by the First Affiliated Hospital with Nanjing Medical University, China. A minimum of 20% tumor content was required for formalin-fixed paraffin-embedded (FFPE) specimens, from which genomic DNA was extracted using QIAamp DNA FFPE Tissue Kit (QIAGEN) following the manufacturer’s instructions. Plasma was extracted from 5 to 10 ml peripheral blood collected in EDTA-coated tubes within 2 h from the blood withdrawn. Circulating tumor DNA (ctDNA) was extracted from the plasma using QIAamp Circulating Nucleic Acid Kit (QIAGEN). Genomic DNA of the whole blood was extracted with DNeasy Blood & Tissue Kit (QIAGEN) as germline control. The DNA quantity was measured on Qubit 3.0 with dsDNA HS Assay Kit (Life Technologies).
Library preparation and sequencing
Sequencing libraries were prepared with KAPA Hyper Prep Kit (KAPA Biosystems) as per manufacturer’s instructions with optimized protocols as previously described [20]. Briefly, fragmented DNA was subjected to end-repairing, A-tailing, indexed-adapter ligation, size selection, and PCR amplification. For targeted enrichment, indexed DNA libraries were pooled together for hybridization with customized xGen lockdown probes (Integrated DNA Technologies) for 416 predefined cancer-relevant genes. Enriched libraries were amplified and subjected for NGS on Illumina Hiseq4000 platforms (Illumina) to a targeted mean coverage depth of 500× for FFPE samples or 3000× for ctDNA samples.
Data processing
After demultiplexing, FASTQ files were processed with Trimmomatic [21] for quality control. Reads were then mapped to human reference genome 19 (hg19) using Burrows-Wheeler Aligner (BWA) [22]. Local realignment and base quality score recalibration were conducted with Genome Analysis Toolkit (GATK) [23]. Mapping rate for each sample is over 99.5% with 82% on-target rate and 99.2% of uniformity (percentage of bases over 0.2× mean coverage depth). SNV and indel mutation calling was performed using VarScan (https://dkoboldt.github.io/varscan) (< 10% minor allele frequency [MAF]) and GATK (> 10% MAF). Identified Mutations were first filtered with dbSNP and 1000 Genome data sets to remove common SNPs. Mutations identified within the whole blood controls were subtracted to exclude germline mutations where applicable. Structural variants were detected using FACTERA with default parameters [24]. ADTEx (https://adtex.sourceforge.net) was used to identify copy number variations (CNVs) with default parameters. All the genetic alterations identified were manually reviewed on Integrative Genomics Viewer (IGV) software for curation [25].
Statistical analysis
Cox proportional hazards model was applied to assess if patients’ baseline characteristics show significant association to the PFS. To each gene where applicable, Wilcoxon ranked sum test was applied to compare the PFS of patients with or without mutations in the specified gene. Gehan-Breslow-Wilcoxon method was used to further assess the PFS curves of different patient groups based on their genotype. P value less than 0.05 was considered statistically significant.
Results
Mutation profiling in Chinese mCRC patients
All patients enrolled in this study were pre-screened as KRAS G12 and G13 mutation-negative using amplification-refractory mutation system (ARMS) [26] analysis based on the NCCN guideline that patients with mutations in KRAS are not suggested for cetuximab-based therapy [6]. The clinical information of the 33 Chinese mCRC patients enrolled in this study was summarized in Table 1 and the detailed clinical characteristics of each patient were listed in Additional file 1: Table S1. Pre-cetuximab-based treatment, tumor samples from 31 patients and plasma samples from 2 patients (Patient 4 and Patient 33) were analyzed using NGS targeting 416 cancer-relevant genes [20]. All patients received at least one course of cetuximab-based therapy until PD was observed. Drug response was evaluated at week 12 post-treatment, and PFS was calculated from the first day of the treatment until PD (Fig. 1). Other than the ECOG score (P = 0.007), no baseline characteristics are correlated to PFS according to Cox proportional hazards model (Table 1).
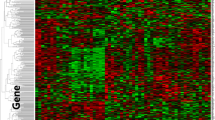
Genetic alterations detected from 33 Chinese mCRC patients using targeted NGS with a 416 cancer-related gene panel. Twenty one genes with at least 4 recurrences are shown in the figure. PFS upon receiving cetuximab are shown on the top. Different mutation types are colour coded: cyan represents single nucleotide variant (SNV), orange represents copy number variation (CNV), and red represents insert/deletion (Indel). Patients received previous chemotherapy are labeled differently as indicated in their PFS
A total number of 411 somatic alterations including single nucleotide variations (SNVs) (n = 320), CNVs (n = 38), and insertion/deletions (indels) (n = 53) distributed in 190 genes were detected in these patients (Fig. 1 and Additional file 2: Table S2). The most frequently mutated gene was TP53 (75.8%), followed by APC (54.5%), SMAD4 (24.2%), and PIK3CA (21.2%). Compared to the published results from The Cancer Genome Atlas (TCGA) [4], the mutation frequency of TP53 was higher in our Chinese mCRC cohort (58.6% in TCGA), while only one FBXW7 mutation (3%) was found in our data set (Additional file 2: Table S2) versus 11.4% in TCGA [4]. Surprisingly, although the 33 patients were pre-screened as KRAS G12 and G13 mutation-negative by ARMS method, we were able to identify 9 out the 33 patients with KRAS alterations (27.3%) using NGS, including exon 2 (G12 V, G12D and G13C), exon 3 (Q61R), and exon 4 (K117 and A146) mutations, as well as 2 cases with KRAS amplifications (Additional file 3: Figure S1 and Additional file 2: Table S2).
Identification of SMAD4 and NF1 mutations as biomarkers for poor prognosis upon cetuximab-based therapy
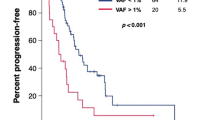
As shown in Fig. 1, the PFS of all the patients range from 12 days to 959 days, with the median of 211 days. To further identify potential prognosis biomarkers for cetuximab-based therapy, we performed Wilcoxon ranked sum test [27] by comparing the PFS of patients divided by the mutation status in a specific gene. Of the 21 top mutated genes in Fig. 1, significant difference of PFS from the mutated group and the corresponding wildtype group was discovered to be caused by two genes, SMAD4 (P = 0.026) and NF1 (P = 0.034) (Additional file 4: Table S3). The median PFS in the SMAD4 mutated (SMAD4mut) or NF1 mutated (NF1mut) subsets was 90 days and 53.5 days compared to the 250 days and 211 days in the corresponding wildtype group, respectively (Table 2). Of note, two patients carrying both SMAD4 and NF1 mutations had the shortest PFS in our data set (12 and 37 days, respectively, Fig. 1). Furthermore, significance was also found in PFS curves between SMAD4mut and SMAD4wt groups (P = 0.0081) (Fig. 2a), and that of NF1mut and NF1wt groups (P = 0.0028) (Fig. 2b). In addition, when combining patients carrying mutated SMAD4 and/or NF1 (SMAD4mut or NF1mut), the difference was still significant comparing to patients carrying both wildtype SMAD4 and NF1 (SMAD4wt and NF1wt) (P = 0.0063) (Fig. 2c), suggesting that at least one mutated SMAD4 or NF1 could serve as potential biomarker to predict poor cetuximab prognosis. Furthermore, according to the drug response evaluation at week 12 post-ceuximab-based treatment, all the SMAD4mut or NF1mut patients showed PD or stable disease (SD), which is considered as no response according to the Guidance for Industry Clinical Trial Endpoints for the Approval of Cancer Drugs and Biologics (Fig. 1) [19].
Comparison of PFS curves according to the mutation status of a defined gene or gene combination upon receiving cetuximab-based therapy. a. Comparison of PFS curves of the SMAD4mut group and the SMAD4wt group. b. Comparison of PFS curves of the NF1mut group and the NF1wt group. c. Comparison of PFS curves of the SMAD4mut or NF1mut group comparing with the SMAD4wt and NF1wt group. The P value is calculated according to Gehan-Breslow-Wilcoxon test. Figures are made using GraphPad Prism 5
However, no difference in PFS was observed in the KRASmut group comparing to the KRASwt group in this study (Additional file 3: Figure S1). Of note, the two patients with KRAS amplification showed a trend of shorter PFS (53 days and 99 days, respectively) with no response to EGFR blockade, in accordance with the results described by a previous study [28].
In summary, analysis of the mutation profiles in the 33 mCRC patients in combination with the efficacy of their cetuximab-based therapy demonstrated that SMAD4 or NF1 mutations are promising potential biomarkers for cetuximab-based therapy in Chinese mCRC patients.
Discussion
In this study, we analyzed the mutation landscape of 33 Chinese mCRC specimens using NGS targeting 416 cancer-relevant genes. Most frequently mutated genes are TP53 (75.8%), APC (54.5%), SMAD4 (24.2%), and PIK3CA (18.2%). In comparison with data from TCGA [4], TP53 was mutated more frequently in the Chinese mCRC patients, while the mutation frequency of FBXW7 was much lower. Furthermore, we have identified SMAD4 and NF1 as candidates for potential prognosis biomarkers for cetuximab-based therapy, as patients carrying mutated SMAD4 and/or NF1 genes had significantly shorter PFS than the SMAD4wt and NF1wt group.
SMAD4 plays an important role as a common mediator in the transcriptional regulator complex in the TGF-β pathway [29], which is a bypass signaling pathway of EGFR-mediated signaling pathway for cell proliferation, differentiation and survival. It has been shown that loss of SMAD4 expression is associated to poor overall survival and poor prognosis of chemotherapy in CRC patients [30, 31]. Furthermore, studies of SMAD4 as potential biomarkers for cetuximab prognosis have been reported previously, although insufficient and controversial. Two groups conclude that alterations in SMAD4 are associated with a poor cetuximab prognosis [17, 32], while another study made the opposite conclusion that SMAD4 is a biomarker for superior cetuximab prognosis [33]. Our results are in agreement with the former two studies that SMAD4 is likely correlated to primary resistance to anti-EGFR therapy.
NF1 encodes neurofibromin 1, which functions as a negative regulator of the RAS signaling pathway downstream of EGFR [34]. To our knowledge, our study is the first report of NF1 as a biomarker for the prognosis of cetuximab-based therapy in mCRC. It is likely that deregulation of the TGF-β and RAS pathways due to mutations in SMAD4 and NF1 results in primary resistance to anti-EGFR blockade, possibly via the activation of the bypass or downstream signalling pathways of EGFR.
Although these patients were screened as KRAS G12 and G13 mutation-negative by ARMS method, we identified 9 out of 33 patients with KRAS alterations using NGS. By comparing the KRASmut and KRASwt patients, no difference was seen in PFS curves (P = 0.2826), which is not consistent with the current treatment guideline that patients carrying KRAS mutations, especially exon 2 mutations, present primary-resistance to anti-EGFR blockade [6]. The reason for the inconsistency might be due to the relative smaller sample size in this study. In addition, it has been reported in several studies that patients harbouring KRAS G13D mutation, comparing to other exon 2 mutations, might benefit from cetuximab-based therapy [35,36,37]. Mutations in other exons have different effects on cetuximab efficacy. Studies show that KRAS Q61 mutation has adverse effect on cetuximab prognosis while A146 seems to have little effect [16]. In our patient cohort, the KRAS mutations were evenly distributed in three exons at 6 spots (Additional file 3: Figure S1) after pre-screening for KRAS exon 2 mutation-negative, which might put bias on patient selection and thus makes the difference of PFS curves of KRASmut and KRASwt not significant.
Conclusion
In this study, we have depicted pan-cancer mutation profiles from 33 Chinese mCRC patients. We further identified SMAD4 and NF1 mutations as potential biomarkers for poor prognosis of cetuximab-based therapy, which needs to be further validated in a larger patient cohort.
Abbreviations
- ARMS:
-
Amplification refractory mutation system
- CNV:
-
Copy number variation
- CR:
-
Complete response
- ctDNA:
-
Circulating tumor DNA
- EGFR:
-
Epidermal growth factor receptor
- FFPE:
-
Formalin-fixed paraffin-embedded
- GATK:
-
Genome Analysis Toolkit
- hg19:
-
Human reference genome 19
- IGV:
-
Integrative Genomics Viewer
- indel:
-
Insertion/deletion
- MAF:
-
Minor allele frequency
- mCRC:
-
Metastatic colorectal cancer
- NCCN:
-
National Comprehensive Cancer Network
- NGS:
-
Next-generation sequencing
- PD:
-
Progressive disease
- PFS:
-
Progression-free survival
- PR:
-
Partial response
- RECIST:
-
Response Evaluation Criteria In Solid Tumors
- SD:
-
Stable disease
- SNV:
-
Single nucleotide variation
- TCGA:
-
The Cancer Genome Atlas
References
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32.
Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7.
Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–13.
Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–9.
Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18(20):3098–103.
Colinical Practice Guidelines in Oncology (NCCN Guidelines) Colon Cancer. 2017:COL-1-11.
Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14(4):235–46.
Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pinter T, Lim R, Bodoky G, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360(14):1408–17.
Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(10):1626–34.
Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351(4):337–45.
Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, Ciardiello F. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6(9):519–27.
Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757–65.
Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. Extended RAS Mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Annals of oncology : official journal of the European Society for Medical Oncology. 2015;26(1):13–21.
Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69(5):1851–7.
Sood A, McClain D, Maitra R, Basu-Mallick A, Seetharam R, Kaubisch A, Rajdev L, Mariadason JM, Tanaka K, Goel S. PTEN gene expression and mutations in the PIK3CA gene as predictors of clinical benefit to anti-epidermal growth factor receptor antibody therapy in patients with KRAS wild-type metastatic colorectal cancer. Clin Colorectal Cancer. 2012;11(2):143–50.
De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. The Lancet Oncology. 2010;11(8):753–62.
Sarshekeh AM, Advani S, Overman MJ, Manyam G, Kee BK, Fogelman DR, Dasari A, Raghav K, Vilar E, Manuel S, et al. Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS One. 2017;12(3):e0173345.
Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92(3):205–16.
Guidance for Industry Clinical Trial Endpoints for the Approval of Cancer Drugs and Biologics Edited by U.S. Department of Health and Human Services, Food and Drug Administration. Center for Drug Evaluation and Research. CDER: Center for Biologics Evaluation and Research (CBER); 2007. p. 7.
Jin Y, Shao Y, Shi X, Lou G, Zhang Y, Wu X, Tong X, Yu X. Mutational profiling of non-small-cell lung cancer patients resistant to first-generation EGFR tyrosine kinase inhibitors using next generation sequencing. Oncotarget. 2016;7(38):61755–63.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20.
Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–60.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8.
Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M, Alizadeh AA. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30(23):3390–3.
Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–92.
Little S: Current Protocols in Human Genetics; 1995.
Wilcoxon F. Individual comparisons by ranking methods. Biom Bull. 1945;1(6):80–3.
Mekenkamp LJ, Tol J, Dijkstra JR, de Krijger I, Vink-Borger ME, van Vliet S, Teerenstra S, Kamping E, Verwiel E, Koopman M, et al. Beyond KRAS mutation status: influence of KRAS copy number status and microRNAs on clinical outcome to cetuximab in metastatic colorectal cancer patients. BMC Cancer. 2012;12:292.
Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov. 2012;11(10):790–811.
Kozak MM, von Eyben R, Pai J, Vossler SR, Limaye M, Jayachandran P, Anderson EM, Shaffer JL, Longacre T, Pai RK, et al. Smad4 inactivation predicts for worse prognosis and response to fluorouracil-based treatment in colorectal cancer. J Clin Pathol. 2015;68(5):341–5.
Alazzouzi H, Alhopuro P, Salovaara R, Sammalkorpi H, Jarvinen H, Mecklin JP, Hemminki A, Schwartz S Jr, Aaltonen LA, Arango D. SMAD4 as a prognostic marker in colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(7):2606–11.
Lupini L, Bassi C, Mlcochova J, Musa G, Russo M, Vychytilova-Faltejskova P, Svoboda M, Sabbioni S, Nemecek R, Slaby O, et al. Prediction of response to anti-EGFR antibody-based therapies by multigene sequencing in colorectal cancer patients. BMC Cancer. 2015;15:808.
Yu J, Wu WK, Li X, He J, Li XX, Ng SS, Yu C, Gao Z, Yang J, Li M, et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut. 2015;64(4):636–45.
Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104(4):593–604.
De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304(16):1812–20.
Mao C, Huang YF, Yang ZY, Zheng DY, Chen JZ, Tang JL. KRAS p.G13D mutation and codon 12 mutations are not created equal in predicting clinical outcomes of cetuximab in metastatic colorectal cancer: a systematic review and meta-analysis. Cancer. 2013;119(4):714–21.
Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol. 2012;30(29):3570–7.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 91429308 and 81572389), and the Science and Technology Development Foundation of Nanjing Medical University (grant number 2016NJMU064).
Availability of data and materials
Raw NGS data cannot be made publicly available for patients’ confidentiality.
Author information
Authors and Affiliations
Contributions
ZM, YWS, YS, ZF, and YG made substantial contributions to the concept and design of the study; ZM, PL, XC, BW, YX, DZ, YS, ZF and YG were involved in the acquisition of patients’ sample, clinical information and patient follow-up; YD and XM performed the NGS testing; ZM, YWS, YD, XM, XW made substantial contributions to the data analysis and interpretation; ZM, YWS, and YG drafted the manuscript. All authors read and approved the final manuscript to be published.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the ethic committee of the First Affiliated Hospital with Nanjing Medical University. All the patients enrolled in this study have provided written informed consents for specimen collection, genetic testing, and participation in the research anonymously.
Consent for publication
Not applicable.
Competing interests
Yang W. Shao, Yan Ding, Xiangyuan Ma, and Xue Wu are the shareholders or employees of Geneseeq Technology Inc.; Yewei Xia, and Dongqin Zhu are the employee of Nanjing Geneseeq Technology Inc..
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Clinical characteristics of the 33 Chinese mCRC patients enrolled in this study. (XLSX 12 kb)
Additional file 2:
Table S2. Genetic alterations detected in targeted NGS with a 416 pan-cancer gene panel from the 33 Chinese mCRC patients. (XLSX 53 kb)
Additional file 3:
Figure S1. Prognosis of cetuximab-based treatment between KRASmut and KRASwt patients. A. Mutation site and type of the 9 KRAS mutations or genetic alterations detected in this study associated with PFS and drug response to cetuximab-based therapy. KRASwt indicates the median PFS of wildtype KRAS patients (230.5 days). The minor allele frequency (MAF) of each KRAS mutation is displayed in red. SMAD4 and NF1 mutations are shown in yellow in the Fig. B. PFS curves of patients with (KRASmut) and without (KRASwt) mutations in KRAS. P value is calculated according to Gehan-Breslow-Wilcoxon test. Figure is made using GraphPad Prism 5. (JPEG 983 kb)
Additional file 4:
Table S3. Statistical difference of PFS of patients with or without mutations in a specific gene. (XLSX 10 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Mei, Z., Shao, Y.W., Lin, P. et al. SMAD4 and NF1 mutations as potential biomarkers for poor prognosis to cetuximab-based therapy in Chinese metastatic colorectal cancer patients. BMC Cancer 18, 479 (2018). https://doi.org/10.1186/s12885-018-4298-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-018-4298-5