Abstract
Background
An early sepsis-induced acute kidney injury (sepsis-AKI) biomarker is currently in needed. Urinary neutrophil gelatinase-associated lipocalin (uNGAL) is a candidate of sepsis-AKI biomarker but with different cut-point values. Urinary exosomal activating transcriptional factor 3 (uATF3) has been mentioned as an interesting biomarker.
Methods
We conducted experiments in mice and a prospective, multicenter study in patients as a proof of concept that urine exosome is an interesting biomarker. An early expression of ATF3 in kidney of CD-1 mice at 6 h after cecal ligation and puncture implied the possibility of uATF3 as an early sepsis-AKI biomarker. Increase serum creatinine (Scr) ≥0.3 mg/dL from the baseline was used as an AKI diagnosis and urine was analyzed for uATF3 and uNGAL. Patients with baseline Scr at admission ≥1.5 mg/dL were excluded.
Results
The analysis showed higher Scr, uNGAL and uATF3 in patients with sepsis-AKI in comparison with patients with sepsis-non-AKI and healthy volunteers. A fair correlation, r2 = 0.47, between uATF3 and uNGAL was showed in sepsis-AKI group with Scr ≥2 mg/dL. To see if uATF3 could be an early sepsis-AKI biomarker, urine sample was collected daily during the first week of the admission. In sepsis-AKI and sepsis-non-AKI groups, uNGAL were 367 ± 43 ng/mL and 183 ± 23 ng/mL, respectively; and uATF3 were 19 ± 4 ng/mL and 1.4 ± 0.8 ng/mL, respectively. With the mean value of uNGAL and uATF3 in sepsis AKI as a cut-off level, AUROC of uNGAL and uATF3 were 64% (95% CI 0.54 to 0.74) and 84% (95% CI 0.77 to 0.91), respectively.
Conclusions
Urine exosome is an interesting source of urine biomarker and uATF3 is an interesting sepsis-AKI biomarker.
Similar content being viewed by others
Background
Sepsis is the systemic inflammatory response syndrome (SIRS) in the response of infectious processes regardless of organisms [1]. Sepsis is a world-wide health care problem and a leading cause of death of patients in intensive care unit [2]. On the other hand, acute kidney injury (AKI) is a clinical syndrome of an abrupt renal dysfunction leading to uremic toxins accumulation and several organs dysfunction [3]. It is interesting that more than 50% of patients with sepsis develop AKI and sepsis patients with AKI had a higher mortality rate [4, 5]. The definition of sepsis-induced AKI (sepsis-AKI) is an AKI occurs simultaneously or subsequently after sepsis with the exclusion of other causes of AKI [2]. Despite an advance in an understanding of pathogenesis and sepsis-AKI therapy in animal models, the translational research into a human condition is currently unsuccessful. This, at least in part, demonstrates the requirement for early sepsis-AKI biomarkers [6]. Serum creatinine (Scr), a current standard biomarker of AKI, is a late renal injury biomarker which does not a good representative of sepsis-AKI [7–10]. Thus, several potential biomarkers for detecting sepsis-AKI have been studies. Urinary neutrophil gelatinase associated lipocalin (uNGAL) is one of the promising sepsis-AKI biomarker, but still have a debate on the cut-off value [11, 12]. The difference cut-off value of uNGAL might due to the possibility of different sources of neutrophil gelatinase-associated lipocalin (NGAL), such as neutrophil and other internal organs, etc., especially, in patients with sepsis [13, 14].
Exosomes are the nano-vesicles released by most of the cells as a method of cell communication for either normal physiologies or many pathological processes [15]. These nano-vesicles are some parts of cell membrane containing several proteins including the genetic materials [16, 17]. In another word, exosome is a membrane-protected source of biological materials. Hence, urine exosome is an interesting source of urine biomarkers [17–19]. Activating transcription factor 3 (ATF3) is one of the transcriptional factor protected inside urine exosome which expressed in urine of AKI in patients and animal models [20, 21]. Renal ATF3 is activated in mouse models of ischemic reperfusion injury and cisplatin induced nephrotoxicity [22, 23]. Interestingly, ATF3 downstream functions are anti-apoptosis and anti-inflammation through the attenuation of pro-inflammatory mediators and the alteration of the chromatin binding site in transcriptional factors [21, 22, 24, 25]. Hence, ATF3 should be involved in the pathophysiology of sepsis, AKI and sepsis-AKI. Unfortunately, ATF3 detection in total urine and non-exosomal urine fraction is inconsistent [20, 21] due, in part, to the degradation of transcriptional factor in urine [26]. But ATF3 seems to be protected inside exosome membrane and allow easier detection [20]. Therefore, we hypothesized that urinary exosomal ATF3 (uATF3) should be an interesting biomarker of sepsis-AKI.
Materials and methods
Cecal ligation and puncture sepsis model and kidney immunohistochemistry
The US National Institutes of Health (NIH) criteria and protocols were followed. Male, 8–10 week old CD-1 mice (National Laboratory Animal Center, Nakhon Pathom, Thailand) were used. Cecal ligation and puncture sepsis model (CLP) was performed under isoflurane anesthesia as previously details [10, 27]. In brief, cecum was ligated at 12 mm from the cecal tip, punctured with a 21-gauge needle and squeezed for a small amount of feces. Supplementary fluid, 1 mL of normal saline, was administered at the post-operational period. However, antibiotic was not used because the effect of antibiotic to urinary exosome was uncertain. Only the abdominal incision for cecal identification was done in sham group. Blood (50 μL) was collected through tail vein nicking for the time-point analysis of mouse serum NGAL (sNGAL) (R&D systems, MN, USA) and serum creatinine (Scr) (QuantiChrom Creatinine assay DICT-500, Hayward, CA, USA). Mice were euthanized under isoflurane anesthesia. Kidney, liver and spleen were put in liquid nitrogen then kept at −80 °C until used. Kidney was also fixed in 10% neutral buffered formalin and embedded in paraffin for the immunohistochemical section of 45 μm thickness with anti-mouse NGAL (R&D systems, MN, USA) and anti-mouse ATF3 (Santa cruz, CA, USA) as previously described [27]. Western Blot analysis of tissue ATF3 and activated caspase-3 (Cell Signaling, MA, USA) were also performed. The protocol for the experiment was approved by the Animal Experimentation Ethics Committee of Chulalongkorn University.
Patient selections
Spot urine collection was performed during January 2014 to August 2014 at the King Chulalongkorn Memorial Hospital (KCMH) and three other general hospitals. To include patients with documented early sepsis-AKI during the hospitalization, all adult patients (age more than 18-year-old) in internal medicine ward with sepsis by the demonstration of systemic inflammatory response syndrome (SIRS) with the evidence or the suspicious of infectious causes [1]. Additionally, AKI defined as Scr increase ≥0.3 mg/dL from baseline followed KDIGO and AKIN criteria [3]. Then sepsis-induced AKI (sepsis-AKI) defined as sepsis with Scr increase ≥0.3 mg/dL from baseline [3]. Otherwise, patients with sepsis but Scr increase ≤0.3 mg/dL were classified as sepsis-non-AKI. Exclusion criteria were (i) patients with the baseline Scr (first Scr measured) higher than 1.5 mg/dL or other history of chronic kidney diseases, (ii) pregnant women, (iii) chronic systemic diseases with proteinuria such as diabetic nephropathy, lupus nephritis, etc.; (iv) obstructive uropathy, and (v) post kidney transplantation. Baseline Scr were retrospectively reviewed from the medical records of at least 3-month earlier from the participant hospitals. Patients with sepsis with elevated Scr at the time of admission was firstly determined as sepsis induced acute kidney injury unless they met the exclusion criteria (1st time Scr >1.5 mg/dL). Renal sonogram was used to exclude the preexisting chronic kidney disease (CKD) in some patients. Then the participants were followed with daily first morning spot urine and serum samples collection during the 1st week of admission period.
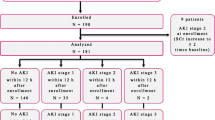
Of all 382 patients, most of the patients had underlying diseases and only 139 patients were included and approximately half of these patients were sepsis-AKI (n = 79) (Fig. 1 and Table 1) implied the importance of AKI in sepsis. Then 8 patients with sepsis-AKI were randomly selected by propensity score matched in gender, age and comorbidity to compare with 8 healthy volunteers group served as discovery set. Subsequently, all urine samples from patients who underwent the new onset of sepsis-AKI (n = 79) during the 1st week of admission were analyzed in comparison with sepsis-non-AKI (n = 60). Of note, there were 17 and 11 patients of sepsis-AKI and non sepsis-AKI, respectively, transferred to intensive care unit. For the urine collection, urine was centrifuged at 1,000x g for 10 min to remove debris, and stored at −80 °C until used.
This study was approved by the Institutional Review Board of Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand and adhered to ARRIVE guideline/methodology.
Measurement of serum creatinine, urine NGAL and urinary exosomal ATF3 of patients’ sample
uNGAL and Scr were measured by ELISA assay (R&D systems, MN, USA) and enzymatic based automated analyzer, respectively. Urinary exosomal ATF3 was measured by Western blot analysis with the quantitative analysis by the calculation from recombinant ATF3 (Santa cruz, CA, USA). Urine exosomes were isolated by the differential centrifugation method as described previously [20]. In brief, frozen urine was thawed and 10 mL of urine was centrifuged at 17,000x g for 15 min. Then the supernatant from the first spin was centrifuged at 200,000x g for 1 h and suspended by 50 μL of 1% SDS in 50 mM Tris. The analysis of Tumor susceptible gene 101 (TSG101), one of the exosome biomarkers [28], was used to support the efficiency of this method. The initial urinary exosome-associated proteins (50 μL) were loaded in different volume into each well depended on urine creatinine (Ucr). This method of exosome biomarker normalization by Ucr was also described previously [20, 29]. The quantitative values of ATF3 by Western blot analysis were calculated from the density of 3 known concentrations of recombinant ATF3 (MyBiosource, CA, USA) loaded in every gel along with the samples.
Western blot analysis
Sample protein were separated by 1day sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis and transferred into a nitrocellulose membrane under Towbin transfer buffer (TBS). The membranes were blocked with 5% milk in TBS plus 0.1% tween-20 for 1 h at room temperature then probed overnight at 4 °C with anti-mouse ATF3 and anti-mouse activated caspase-3 (Cell Signaling, MA, USA) for mouse internal organs or with anti-human ATF3 (Santa cruz, CA, USA) and anti-TSG101 (Cell Signaling, MA, USA) for urine exosome samples. To see if there is TSG101 in the non-exosome urine fraction after centrifuged at 200,000x g (soup), acetone precipitation was used to concentrate the soluble protein in soup fraction before Western blot analysis in parallel with urine exosome fraction (Fig. 4a). For the visualization, the blot was incubated with appropriate horseradish peroxidase-linked secondary antibodies (1:5,000) (Jackson ImmunoResearch Laboratories, PA, USA) and visualized for the chemiluminescence with SuperSignal® (Thermo Scientific, IL, USA) and C-DiGit® Blot Scanner (Li-Cor BioTech, NE, USA).
Statistical analysis
Data was demonstrated as mean ± SD and the differences between groups were examined for statistical significance by unpaired student t-test or one-way analysis of variance (ANOVA) with Tukey’s comparison test in the analysis of experiments with 2 and 3 groups, respectively. Time-point data was examined by two-way ANOVA with Bonferroni post hoc analysis. Survival analysis was performed by log-rank test. P value <0.05 was considered to be a statistically significant. Area under receiver operating characteristic (AUROC), sensitivity and specificity were calculated in relative to increased Scr ≥0.3 mg/dL from baseline as the sepsis-AKI diagnosis. The logistic regression model was fitted to the data to determine the influence of uNGAL and uATF3 and also adjusted by those clinical variables (age, comorbidity, APACHE II score) that were shown to have influence on the risk of AKI. The ability of these models to predict AKI was analyzed by the Hosmer–Lemeshow goodness-of-fit test. Then, the discriminative ability of uNGAL and uATF3 for AKI diagnosis was determined by the AUROC. In addition, the net reclassification improvement (NRI) and the category-free NRI (cfNRI) were performed to demonstrate the additive value of preexisting risk prediction model. NRI demonstrates the quality of predicted probabilities as a consequence of adding a new marker to the existing model. Statistical analysis was performed using SPSS for Windows 15.0 (SPSS for Windows; Chicago, IL) and GraphPad Prism (version 6.00; GraphPad Software, La Jolla, CA) software. Reclassification analyses were performed in R using the regression modelling strategies (rms) package for calculating NRI and cfNRI.
Results
Both NGAL and ATF3 expressed early in kidney of sepsis mice
Lethal sepsis model of cecal ligation and puncture (CLP) with survival rate at 25% (Fig. 2a) was used. Although Scr did not significantly increase in the first 6 h after CLP, mice clinical symptoms of illness and serum NGAL (sNGAL) were observed at 6 h of the surgery (Fig. 2b) consistent with previous publications [10, 27]. At 18 h both Scr and sNGAL were higher than the baseline (Fig. 2b). Because at 6 h-post CLP was the earliest time-point of increased sNGAL, kidney histology at 6 h after CLP was analyzed for NGAL and ATF3 as early-sepsis biomarker candidates. Indeed, NGAL expressed in cytoplasm and ATF3 expressed in nuclei of the renal tubular cell (Fig. 2c, d) at 6 h after CLP but not in sham group (data not showed). Moreover, the activated caspase-3 also detectable in kidney at 6 h of CLP by Western blot analysis (Fig. 2e, f). In addition, ATF3 and NGAL expressed in kidney, liver and spleen at 6 h of CLP sepsis mice (Fig. 3) but not in sham (showed only kidney). Due to an early anuria in sepsis mice, we could not analyze urine biomarker in this model.
Survival analysis of the cecal ligation and puncture (CLP) model used in the study (a) and time-course of serum neutrophil gelatinase associated lipocalin (sNGAL) and serum creatinine (Scr) (b) were demonstrated. The representative pictures of the immunohistochemistry of mouse kidney after 6 h of CLP sepsis surgery for NGAL (c) and activating transcriptional factor 3 (ATF3) (d). NGAL and ATF3 were demonstrated in cytoplasm/brush border and nuclei of cortical renal tubules cells, respectively, and both parameters could not be detected in kidney of sham surgery (data not showed). Additionally, activated caspase-3 was also analyzed in kidney sample at 6 h of CLP and sham surgery (e). The comparison of Caspase-3 Western Blot density between kidney of sham and CLP group measured from C-DiGit® Blot Scanner was also showed (f)
The Western blot analysis of ATF3 in different organs of mice at 6 h after cecal ligation and puncture sepsis surgery (upper panel) and the density scoring (lower panel) (n = 3-5/group). The reciprocal organs of mice at 6 h after sham surgery were undetectable (data showed only kidney of sham group)
High urinary NGAL (uNGAL) and urinary exosomal ATF3 (uATF3) in an initial analysis of patients with sepsis-induced acute kidney injury (Sepsis-AKI)
The presentation of exosome in urine with extraction procedures was demonstrated by TSG101 (Fig. 4a). By using propensity score matched in gender, age and comorbidity, 8 from 79 patients with sepsis-AKI were randomly selected for the analysis. Scr of patients with sepsis-AKI and healthy volunteers were 2.15 ± 0.22 and 0.97 ± 0.02 mg/dL, respectively (p <0.001) (Fig. 4b). With simultaneous analysis, uNGAL and uAFT3 of sepsis-AKI versus healthy volunteers were 1,909 ± 838 versus 17 ± 4 ng/mL and 16 ± 4 and 0 ng/mL, respectively (Fig. 4c, d). Interestingly, uATF3 was non-detectable in all healthy volunteers and both uATF3 and uNGAL associated with AKI severity by AKIN classification. The median value of uATF3 versus uNGAL for AKIN stage 1, 2 and 3 were 8.5-11.7, 14.2-16.7, and 18.2-22.3 ng/mL versus 121–374, 529–871, and 1,071-1,533 ng/mL, respectively (p <0.001 among each stage). However, the correlation between all uNGAL and uATF3 was limited (r 2 = 0.39) but increased (r 2 = 0.47) with the selection of Scr ≥2 mg/dL. With Scr < 2 mg/dL, there was even lesser correlation (r 2 = 0.26).
TSG101 (MW 49 kDa) was demonstrated in esosome fraction of urine (Exosome) supported the efficiency of urine exosome extraction procedure and TSG101 could not be detected in non-exosome urine fraction (soup) (a). The cross-sectional analysis from sepsis-AKI patients regardless of the onset of sepsis-AKI for serum creatinine (Scr) (b), urinary neutrophil gelatinase associated lipocalin (uNGAL) (c) and urinary activating transcriptional factor 3 (uATF3) (d) were showed (n = 8/group)
Urinary NGAL (uNGAL) and urinary exosomal ATF3 (uATF3) in a 7 days follow-up of patients with sepsis-induced acute kidney injury (Sepsis-AKI) showed a potential as an early sepsis-AKI biomarkers
There were data from 79 and 60 patients with sepsis-AKI and non sepsis-AKI, respectively, available for the analysis. Of note, 10 from 79 patients (12.7%) in sepsis-AKI group were classified as septic shock. Other patients were categorized as severe sepsis due to there was some organ injuries in these patients. Because there was different onset of sepsis-AKI during 1 week of the follow-up, all data of the 7 days follow-up were combined. With the combination data, Scr, uNGAL and uATF3 of sepsis-AKI and sepsis-non-AKI group were 1.25 ± 0.11 mg/dl, 367 ± 43 ng/mL and 19 ± 4 ng/mL and 0.97 ± 0.02 mg/dL, 183 ± 23 ng/mL and 1.4 ± 0.8 ng/mL, respectively (p <0.001 all) (Fig. 5a-c). Of note, most of the patients in this study were non-oligulic sepsis-AKI (68/79 or 86%) (urine output more than 0.5 mL/kg/h) during the observational period. As expected, uNGAL in sepsis-AKI was higher than sepsis non-AKI group reciprocal to previous studies [30]. Interestingly, uATF3 was also higher in sepsis-AKI group. It is interesting that uATF3 was undetectable in nearly all time-points of sepsis non-AKI group implied the high specificity of uATF3 for sepsis-AKI (Fig. 5c). Although both uNGAL and uATF3 correlated with Scr levels, uATF3 provided the better differentiation between sepsis-AKI and sepsis non-AKI (Fig. 5d, e).
To explore an early sepsis-AKI biomarker potential, uNGAL and uATF3 at 1 day before and at 1st and 2nd day of Scr ≥0.3 mg/dL from baseline was demonstrated (Fig. 5d, e, 6a and b). There was a statistical significant difference of 1st day uATF3 following AKI onset compared with baseline (p <0.05) (Fig. 6b). However, uNGAL but not uATF3 showed a tendency of the higher value at 1 day before AKI onset (Fig. 6a). To calculate area under receiver operating characteristic (AUROC) curve and sensitivity/specificity of uNGAL and uATF3 for sepsis-AKI diagnosis, all time-point data of matched value between uNGAL and uATF3 from patients with sepsis-AKI were combined. The gold standard of sepsis-AKI was Scr ≥0.3 mg/dL from baseline and the cut-off value of uNGAL and uATF3 was 188 and 12 ng/mL, respectively, from the mean level of sepsis-non-AKI group. We found that AUROC of uNGAL and uATF3 were 64% and 84%, respectively (Fig. 7). It seems that uATF3 showed a comparable sensitivity but higher specificity for sepsis-AKI diagnosis in comparison to uNGAL. Urine NGAL and uATF3 performance with different cut-off values were demonstrated (Table 2). At 1 day before AKI, the sensitivity of uNGAL and uATF3 were comparable but uATF3 showed higher specificity.
To clearly demonstrated if new biomarkers could be detected at an early stage of sepsis-AKI, values of urinary neutrophil gelatinase associated lipocalin (uNGAL) (a) and urinary activating transcriptional factor 3 (uATF3) (b) at baseline, 1 day before and at 1st and 2nd day of Scr higher than baseline for 0.3 mg/dL was showed. *p <0.05 compared with baseline levels
The area under the receiver operating characteristic (AUROC) curve analysis of sensitivity and specificity of the urinary neutrophil gelatinase associated lipocalin (uNGAL) (a) and urinary activating transcriptional factor 3 (uATF3) (b) with the alteration of Scr higher than 0.3 mg/dL as a gold standard of sepsis-AKI diagnosis
In a multiple logistic regression model, patients with uNGAL at 150 ng/mL had a fivefold risk of AKI (OR, 5.6; 95% CI, 3.1 to 7.4); those patients with uATF3 at 12 ng/mL showed 8-fold higher risk of developing AKI (OR, 8.2; 95% CI, 5.6 to 12.8) (Table 3). Presence of comorbidity was associated with an OR of 2.8 (95% CI, 1.6 to 3.8), and high APACHE score were associated with an OR of 4.2 (95% CI, 1.9 to 6.6). Regarding the risk prediction model, a fair performance was obtained (Hosmer–Lemeshow P value =0.77), with AUC = 0.79 (95% CI, 0.70 to 0.88). To determine the addition values of uNGAL and uATF3 to the clinical model (age, comorbidity and APACHE II score), the category-free NRI (cfNRI) was calculated (Table 4). Urine NGAL, uATF3, and both values in combination enhanced predictive risk in 67%, 69% and 49%, respectively, in sepsis-AKI patients, and 35%, 44% and 52%, respectively, in sepsis-non-AKI patients. In addition, uNGAL, uATF3, and the combined biomarkers improved overall cfNRI in 75%, 83% and 80%, respectively (Table 4).
Discussion
Urinary exosome is an interesting source of urine biomarker and urinary exosomal ATF3 (uATF3) was a candidate of early AKI biomarker [20, 21]. ATF3 and NGAL expressed in kidney histology earlier than Scr of sepsis mice. Indeed, the analysis of urine patients with sepsis-AKI demonstrated higher uNGAL and uATF3 in comparison with either healthy volunteer or non-sepsis AKI. Moreover, only uATF3, but not uNGAL, increased during the early period of sepsis-AKI with the better area under receiver operating characteristic curve (AUROC) over uNGAL.
To see if ATF3 is a good candidate for the early sepsis-AKI biomarker, we explore in a mouse model due to the known sepsis onset in the model but not in patients. Because sNGAL increased as early as 6 h after cecal ligation and puncture (CLP) with the onset of clinical abnormality consistent with previous publications [10, 27, 31], we determined 6 h after CLP as the early phase of sepsis-AKI. Interestingly, both NGAL and ATF3 expressed in renal tubular cell at brush border and nuclei, respectively, suggested different functions of these proteins. Indeed, NGAL produced and excreted out from different cells in response to organs injury to controlling the extracellular iron reactive oxygen species then re-absorbed and secreted through the proximal and distal renal tubular cell, respectively [32]. On the other hand, ATF3 interfered with several intra-nuclei transcriptional factors to inhibit pro-inflammatory cytokine translational processes [22]. Moreover, activated caspase-3 was also detectable from renal tissue at 6 h of CLP by Western blot analysis support the association of apoptosis and ATF3 or NGAL as previously described [33]. Despite ATF3 is not specific to kidney as it was also demonstrated in other organs as previously known [34–36], it seems to be activated only in condition with organs injury as there was no ATF3 expression in sham mice. Then ATF3 in urine should be a better representative renal biomarker than serum ATF3. NGAL, on the other hand, is also not specific to kidney and kidney injury [37] but it could be used as an early-AKI biomarker [14, 38, 39]. Although, the early expression of NGAL and ATF3 in kidney of CLP mice suggested the possibility of early sepsis biomarker, CLP model was anuria. Then we test uNGAL, a more specific renal injury than plasma NGAL, and exosomal uATF3 in urine patient sample.
Indeed, uATF3 and uNGAL were high in patients with sepsis-AKI in comparison with the healthy volunteers but with the poor correlation between both parameters. The correlation is better with the higher Scr. Perhaps, there were different molecular induction pathways of both molecules in the different sepsis-AKI severity. Interestingly, uATF3 was negative in all healthy volunteers implied the high specificity to sepsis-AKI.
For exploration of early sepsis-AKI biomarkers, we collected urine samples of patients with sepsis during the first week of the admission to identify the onset of sepsis-AKI. To show the tendency of biomarkers toward sepsis-AKI, matched data of uNGAL and uATF3 from all time-point were combined. As expected, uATF3 and uNGAL in sepsis-AKI were higher than sepsis-non-AKI group and uATF3 was rarely increased in sepsis-non-AKI. In contrast, there was a baseline level and slightly increase of uNGAL in healthy volunteers and sepsis-non-AKI during the follow-up. This implied an easier decision for the cut-off value of uATF3 than uNGAL. For the clinical application, uNGAL and uATF3 were analyzed in comparison with the day of increased Scr due to the difference onset of sepsis-AKI in these patients. Surprisingly. uNGAL show a tendency to increase at 1 day before increase in Scr, but non-statistically significance, perhaps due to the non-specific response to infection. The loss of the AKI prediction property of uNGAL in our study might due to the very subtle increased in Scr or the non-oliguric septic-AKI in most of the patients. However, our data derived from the very strict criteria of new onset of sepsis-AKI developed during the admission. Nevertheless, only uATF3 increased at the same days of increased Scr. This demonstrated the benefit of uAFT3 over uNGAL in predicting AKI. Despite uATF3 did not increase before Scr, uATF3 but not uNGAL, would be a good additive biomarker for supporting the onset of AKI in sepsis condition with only a subtle increase of Scr. Further studies are needed.
With all matching data of uATF3 and uNGAL with the mean values of sepsis-non AKI group as the cut-off, the sensitivity of uATF3 for sepsis-AKI diagnosis was comparable with uNGAL with the higher specificity (Table 3). Indeed, our data demonstrated the comparable AUROC of uNGAL and proposed uATF3 as another early sepsis-AKI biomarker (Table 5).
In short, our results were a proof of concept that urinary exosome was an interesting source of biomarkers and urinary exosomal ATF3, in particular, was an interesting sepsis-AKI biomarker. More studies will be needed for the proper clinical use of uATF3.
There are several limitations to this study. First, the exosome extraction and analysis method was too complicated for a routine biomarker. The requirement of high urine volume, the normalization by urine creatinine and the Western blot analysis was in-convenient. Second, there were only 14% patients with oliguric sepsis-AKI in our data. Therefore, the levels of uNGAL and uATF3 in oliguric sepsis-AKI might be different. Third, although it was a prospective, cohort study; the number of patients was too small. However, our results could be sufficient to conclude that ATF3 in urine exosome was an interesting sepsis-AKI biomarker that should be supported in the larger studies. Finally, the observation was only a cross-sectional and a short term follow-up data so the association of these biomarkers and clinical outcomes was not explored.
Conclusion
This study identifies ATF3, a transcriptional factor; in urine exosome was an interesting sepsis-AKI biomarker. Since uATF3 could not be detected in most of the patients with sepsis-non-AKI, the qualitative test of uATF3 might be adequate for detecting sepsis-AKI. In contrast, with the baseline of uNGAL, the debates on the cut-off values of uNGAL for sepsis-AKI are still on-going. Our results were also a proof of concept that urine exosomes were an interesting source of urine biomarkers.
Abbreviations
- AKI:
-
Acute kidney injury
- AKIN:
-
Acute Kidney Injury Network
- APACHE II:
-
Acute Physiology and Chronic Health Evaluation II
- ATF3:
-
Activating transcription factor 3
- AUROC:
-
Area under the receiver operating characteristic
- cfNRI:
-
Category-free NRI
- CKD:
-
Chronic kidney disease
- CLP:
-
Cecal ligation and puncture
- KDIGO:
-
Kidney Disease Improving Global Outcomes
- NGAL:
-
Neutrophil gelatinase-associated lipocalin
- Scr:
-
Serum creatinine
- sepsis-AKI:
-
Sepsis-induced acute kidney injury
- SIRS:
-
Systemic inflammatory response syndrome
- uATF3:
-
Urinary exosomal activating transcriptional factor 3
- Ucr:
-
Urine creatinine
- uNGAL:
-
Urinay neutrophil gelatinase-associated lipocalin
References
Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–55.
Goncalves GM, Zamboni DS, Camara NO. The role of innate immunity in septic acute kidney injuries. Shock. 2010;34 Suppl 1:22–6.
Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin Pract. 2012;120:c179–184.
Klenzak J, Himmelfarb J. Sepsis and the kidney. Crit Care Clin. 2005;21:211–22.
Rangel-Frausto MS, Pittet D, Costigan M, Hwang T, Davis CS, Wenzel RP. The natural history of the systemic inflammatory response syndrome (SIRS). A prospective study. JAMA. 1995;273:117–23.
Devarajan P. Emerging biomarkers of acute kidney injury. Contrib Nephrol. 2007;156:203–12.
Shemesh O, Golbetz H, Kriss JP, Myers BD. Limitations of creatinine as a filtration marker in glomerulopathic patients. Kidney Int. 1985;28:830–8.
Stevens LA, Coresh J, Greene T, Levey AS. Assessing kidney function-measured and estimated glomerular filtration rate. N Engl J Med. 2006;354:2473–83.
Doi K, Yuen PS, Eisner C, Hu X, Leelahavanichkul A, Schnermann J, et al. Reduced production of creatinine limits its use as marker of kidney injury in sepsis. J Am Soc Nephrol. 2009;20:1217–21.
Leelahavanichkul A, Souza AC, Street JM, Hsu V, Tsuji T, Doi K, et al. Comparison of serum creatinine and serum cystatin C as biomarkers to detect sepsis-induced acute kidney injury and to predict mortality in CD-1 mice. Am J Physiol Renal Physiol. 2014;307:F939–948.
Di Nardo M, Ficarella A, Ricci Z, Luciano R, Stoppa F, Picardo S, et al. Impact of severe sepsis on serum and urinary biomarkers of acute kidney injury in critically ill children: an observational study. Blood Purif. 2013;35:172–6.
Martensson J, Bell M, Oldner A, Xu S, Venge P, Martling CR. Neutrophil gelatinase-associated lipocalin in adult septic patients with and without acute kidney injury. Intensive Care Med. 2010;36:1333–40.
Supavekin S, Zhang W, Kucherlapati R, Kaskel FJ, Moore LC, Devarajan P. Differential gene expression following early renal ischemia/reperfusion. Kidney Int. 2003;63:1714–24.
Martensson J, Bellomo R. The rise and fall of NGAL in acute kidney injury. Blood Purif. 2014;37:304–10.
Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012;21:R125–134.
Huebner AR, Somparn P, Benjachat T, Leelahavanichkul A, Avihingsanon Y, Fenton RA, et al. Exosomes in urine biomarker discovery. Adv Exp Med Biol. 2015;845:43–58.
Pisitkun T, Johnstone R, Knepper MA. Discovery of urinary biomarkers. Mol Cell Proteomics. 2006;5:1760–71.
Dear JW, Street JM, Bailey MA. Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics. 2013;13:1572–80.
Knepper MA, Pisitkun T. Exosomes in urine: who would have thought…? Kidney Int. 2007;72:1043–5.
Zhou H, Cheruvanky A, Hu X, Matsumoto T, Hiramatsu N, Cho ME, et al. Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int. 2008;74:613–21.
Chen HH, Lai PF, Lan YF, Cheng CF, Zhong WB, Lin YF, et al. Exosomal ATF3 RNA attenuates pro-inflammatory gene MCP-1 transcription in renal ischemia-reperfusion. J Cell Physiol. 2014;229:1202–11.
Cheng CF, Lin H. Acute kidney injury and the potential for ATF3-regulated epigenetic therapy. Toxicol Mech Methods. 2011;21:362–6.
Miller RP, Tadagavadi RK, Ramesh G, Reeves WB. Mechanisms of Cisplatin nephrotoxicity. Toxins (Basel). 2010;2:2490–518.
Li D, Yin X, Zmuda EJ, Wolford CC, Dong X, White MF, et al. The repression of IRS2 gene by ATF3, a stress-inducible gene, contributes to pancreatic beta-cell apoptosis. Diabetes. 2008;57:635–44.
Li HF, Cheng CF, Liao WJ, Lin H, Yang RB. ATF3-mediated epigenetic regulation protects against acute kidney injury. J Am Soc Nephrol. 2010;21:1003–13.
Thomas CE, Sexton W, Benson K, Sutphen R, Koomen J. Urine collection and processing for protein biomarker discovery and quantification. Cancer Epidemiol Biomarkers Prev. 2010;19:953–9.
Leelahavanichkul A, Yasuda H, Doi K, Hu X, Zhou H, Yuen PS, et al. Methyl-2-acetamidoacrylate, an ethyl pyruvate analog, decreases sepsis-induced acute kidney injury in mice. Am J Physiol Renal Physiol. 2008;295:F1825–1835.
Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6(4):287–96.
Zhou H, Yuen PS, Pisitkun T, Gonzales PA, Yasuda H, Dear JW, et al. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69:1471–6.
Fan H, Zhao Y, Zhu JH, Song FC. Urine neutrophil gelatinase-associated lipocalin in septic patients with and without acute kidney injury. Ren Fail. 2014;36:1399–403.
Doi K, Leelahavanichkul A, Hu X, Sidransky KL, Zhou H, Qin Y, et al. Pre-existing renal disease promotes sepsis-induced acute kidney injury and worsens outcome. Kidney Int. 2008;74:1017–25.
Schmidt-Ott KM, Mori K, Li JY, Kalandadze A, Cohen DJ, Devarajan P, et al. Dual action of neutrophil gelatinase-associated lipocalin. J Am Soc Nephrol. 2007;18:407–13.
Cui LY, Yang S, Zhang J. Protective effects of neutrophil gelatinase-associated lipocalin on hypoxia/reoxygenation injury of HK-2 cells. Transplant Proc. 2011;43:3622–7.
Koivisto E, Jurado Acosta A, Moilanen AM, Tokola H, Aro J, Pennanen H, et al. Characterization of the regulatory mechanisms of activating transcription factor 3 by hypertrophic stimuli in rat cardiomyocytes. PLoS One. 2014;9:e105168.
Zhou H, Guo H, Zong J, Dai J, Yuan Y, Bian ZY, et al. ATF3 regulates multiple targets and may play a dual role in cardiac hypertrophy and injury. Int J Cardiol. 2014;174:838–9.
Zhou H, Shen DF, Bian ZY, Zong J, Deng W, Zhang Y, et al. Activating transcription factor 3 deficiency promotes cardiac hypertrophy, dysfunction, and fibrosis induced by pressure overload. PLoS One. 2011;6:e26744.
Otto GP, Hurtado-Oliveros J, Chung HY, Knoll K, Neumann T, Muller HJ, et al. Plasma Neutrophil Gelatinase-Associated Lipocalin Is Primarily Related to Inflammation during Sepsis: A Translational Approach. PLoS One. 2015;10:e0124429.
Haase-Fielitz A, Haase M, Devarajan P. Neutrophil gelatinase-associated lipocalin as a biomarker of acute kidney injury: a critical evaluation of current status. Ann Clin Biochem. 2014;51:335–51.
Ronco C, Legrand M, Goldstein SL, Hur M, Tran N, Howell EC, et al. Neutrophil gelatinase-associated lipocalin: ready for routine clinical use? An international perspective. Blood Purif. 2014;37:271–85.
Su LX, Feng L, Zhang J, Xiao YJ, Jia YH, Yan P, et al. Diagnostic value of urine sTREM-1 for sepsis and relevant acute kidney injuries: a prospective study. Crit Care. 2011;15:R250.
Aydogdua M, Gürsel G, Sancak B, Yeni S, Sarı G, Ta S, et al. The use of plasma and urine neutrophil gelatinase associated lipocalin (NGAL) and Cystatin C in early diagnosis of septic acute kidney injury in critically ill patients. Dis Markers. 2013;34:237–46.
Matsa R, Ashley E, Sharma V, Walden AP, Keating L. Plasma and urine neutrophil gelatinase-associated lipocalin in the diagnosis of new onset acute kidney injury in critically ill patients. Crit Care. 2014;18:R137.
Terzi I, Papaioannou V, Papanas N, Dragoumanis C, Petala A, Theodorou V, et al. Alpha1-microglobulin as an early biomarker of sepsis-associated acute kidney injury: a prospective cohort study. Hippokratia. 2014;18(3):262e8.
Dai X, Zeng Z, Fu C, Zhang S, Cai Y, Chen Z. Diagnostic value of neutrophil gelatinase-associated lipocalin, cystatin C, and soluble triggering receptor expressed on myeloid cells-1 in critically ill patients with sepsis-associated acute kidney injury. Crit Care. 2015;19:223–32.
Nakamura Y, Ishikura H, Nishida T, Kawano Y, Yuge R, Ichiki R, et al. Usefulness of presepsin in the diagnosis of sepsis in patients with or without acute kidney injury. BMC Anesthesiol. 2014;14:88.
Nakamura Y, Murai A, Mizunuma M, Ohta D, Kawano Y, Matsumoto N, et al. Potential use of procalcitonin as biomarker for bacterial sepsis in patients with or without acute kidney injury. J Infect Chemother. 2015;21(4):257e63.
Acknowledgements
We would like to acknowledge the help and support of the Medicine Department physicians and nursing staff at KCMH, Maharat Nakhonratchasima Hospital, Samut Sakhon Hospital, and Nakhon Pathom Hospital. We also thank Dr. Wasan Panyasang, for his helpful advice on statistical analysis.
Funding
This study was funded by grants from Chulalongkorn University (Ratchadapiseksomphot fund from the faculty of medicine and international research integration, Chula Research Scholar, Ratchadaphiseksomphot Endowment Fund) and by National Science and Technology Development Agency (NSTDA: P-13-00505). T.P. was supported by the 90th Year Anniversary of Chulalongkorn University. A.L. also works under the Center of Excellence in Immunology and Immune-mediated Diseases, Department of Microbiology, Chulalongkorn University, Bangkok, Thailand and funded by Ratchadapisaksomphot Fund and the New Faculty Staff Fund from the faculty of medicine, Chulalongkorn University and Research Grant for New Scholars, Thailand Research Fund (TRG5780207).
Availability of data and materials
No data has been submitted to any open access databases. All data supporting the study is presented in the manuscript or available upon request.
Authors’ contributions
TP, WC, AL contributed to conception and outlined the study design. TP, WC, PS, JI, AL conducted the experiments and analyzed the data. TP, WC, PS, JI, NH, AL drafted and wrote the manuscript as well as provided critical review. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable
Ethics approval and Consent to Participate
The animal protocols were approved by the Institutional Animal Care and Use Committee of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand. The human study was conducted in accordance with the declaration of Helsinki and with the approval of the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand. All patients included in the study signed a written informed consent before participating.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Panich, T., Chancharoenthana, W., Somparn, P. et al. Urinary exosomal activating transcriptional factor 3 as the early diagnostic biomarker for sepsis-induced acute kidney injury. BMC Nephrol 18, 10 (2017). https://doi.org/10.1186/s12882-016-0415-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12882-016-0415-3