Abstract
Background
Increased urinary albumin excretion rate is the earliest clinical manifestation of diabetic nephropathy. The development of microalbuminuria in patients with type 1 diabetes mellitus (T1D) usually begins 5 to 15 years after the onset of diabetes. The rate of progression of diabetic nephropathy varies considerably among patients and not always can be explained solely by glycaemic control. The evidence suggests that genetic susceptibility may play a role in the development of diabetes microvascular complications, besides the presence of such risk factors as hyperglycaemia, hypertension, dyslipidaemia and smoking. The aim of the study was to evaluate a link between known genetic risk factors for type 1 diabetes mellitus (HLA-DR3/DR4) and microalbuminuria among patients with the same durations of diabetes.
Methods
Ninety-nine patients with T1D at the age 18–35 years were recruited for the study. The urine albumin excretion rate was normal when <30 mg/24 h; microalbuminuria 30–300 mg/24 h. Genotypes were investigated in 39 patients with normal albumin excretion rate and duration of diabetes 13.46 ± 3.72 years and in 60 patients with microalbuminuria and duration of diabetes 15.28 ± 4.08 years (p = 0.11). Genetic typing of DR3 and DR4 antigens successfully was performed for 99 subjects. Statistical analysis was performed using SPSS v. 20.0.
Results
Genotyping of 99 patients with T1D was performed: no DR3 and DR4 risk alleles were found in 22 (22.22 %) cases, DR3 alleles were present in 47 (47.48 %) cases, DR4 alleles in 25 (25.25 %) cases, and DR3/DR4 alleles in 5 (5.05 %) cases. The highest 24 h albumin excretion rate was found in patients with DRB1 gene expressed DR3 risk alleles group, the lowest - in patients with DRB1 gene with no expression of both DR3 and DR4 antigen. We confirmed the 1.87 (p = 0.021) increased relative risk for microalbuminuria in patients with DR3/DR3 alleles and same duration of diabetes. The distribution of DR3 and DR4 risk alleles was not associated with cardiovascular autonomic neuropathy both in patients with normal albumin excretion rate and microalbuminuria (1.6 vs 2.1; p = 0.21).
Conclusions
The 1.87 (p = 0.021) increased relative risk for microalbuminuria was found in patients with DR3/DR3 alleles and the same duration of diabetes.
Similar content being viewed by others
Background
Type 1 diabetes is a complex disease, the course of which is influenced by many interacting genetic and environmental factors [1]. Glycaemic control and diabetes duration are the most important risk factors for the development of late diabetic complications. However, the rate of progression of nephropathy, retinopathy and neuropathy varies considerably among patients and not always can be explained solely by glycaemic control. Diabetic nephropathy is one of the most threatening complications of T1D, and its rate and severity directly correlates with the duration of T1D [2]. The presence of diabetic nephropathy is related with the greatest risk of premature death in patients with T1D [3]. Patients with diabetic nephropathy are likely to have neurological complications of diabetes including cardiovascular autonomic neuropathy (CAN), the most often overlooked serious complication of diabetes [4]. Therefore, it is very important to detect kidney damage early and to stabilize effectively, because it is reversible in the initial stages [5]. Microalbuminuria is an early sign of diabetic nephropathy. The development of microalbuminuria in patients with T1D usually begins 5 to 15 years after the onset of diabetes and then increases over time, a small proportion of patients develop microalbuminuria within less than 5 years of disease onset [6, 7]. Clinical nephropathy usually occurs after 15–25 years diabetes duration and rarely newly develops when T1D duration is more than 30 years [8].
The mechanisms of development of diabetic nephropathy are not fully clear. The evidence suggests that genetic susceptibility may play a major role in the development of microvascular complications, besides the presence of such risk factors as hyperglycaemia, hypertension, dyslipidaemia and smoking. Based on underlying pathogenesis, polymorphisms of several candidate genes belonging to multiple pathways have been investigated, like the genes related to mechanisms of hyperglycaemia-induced damage (such as advanced glycation end-products and reactive oxygen species increased formation, augmented activity of the aldose reductase pathway); genes related to the renin-angiotensin system; genes coding for cytokines, growth factors and its receptors, glucose transporters; among many others [9].
The Major Histocompatibility Complex (MHC) region on chromosome 6p21 has been robustly replicated as a major risk locus in many immune-mediated diseases, including T1D. Certain haplotypes of human leukocyte antigen (HLA) genes are known to confer the main genetic risk or protection within the MHC region [1]. The major T1D susceptibility locus maps to the class II loci HLA-DRB1 and HLA-DQB1 on chromosome 6p21. The highest risk DR/DQ haplotypes for T1D are DR3-DQA1*0501-DQB1*0201 (DR3) and DR4-DQA1*0301-DQB1*0302 (DR4). Both DR3and DR4 are very strongly associated with T1D: over 93 % of T1D patients have at least one of these alleles, compared to 43 % of controls [10]. Other non-HLA T1D loci in combination have smaller effects on disease risk compared to HLA [11].
The aim of the study was to evaluate a link between known genetic risk factors for type 1 diabetes mellitus (HLA-DR3/DR4) and microalbuminuria.
Methods
Ninety-nine patients with T1D at the age 18–35 years were recruited for the case–control study. The following standard-case definition criteria for T1D were used: diagnosed as diabetic patient by physician; placed on insulin therapy before the 30th birthday; resident of his/her country at the time of the first insulin administration [12]. The anamnesis of microalbuminuria, hypertension renal failure or consumption of such drugs like ACE (angiotensin-converting-enzyme) inhibitors or ARB (aldosterone receptor blockers) was the exclusion criteria in our study. The information about patients was received from endocrinologists and general practitioners of Kaunas County.
The evaluation 24 h urine albumin excretion rate (AER) was based on C. Mogensen [13]: normal AER <30 mg/24 h; microalbuminuria - AER 30–300 mg/24 h, macroalbuminuria - AER >300 mg/24 h. Subjects with macroalbuminuria, - AER >300 mg/24 h were excluded from investigation. Genotypes were investigated in 39 patients with normal AER and duration of T1D 13.46 ± 3.72 years (cases) and in 60 patients with microalbuminuria and duration of diabetes 15.28 ± 4.08 years (controls) (p = 0.11). Genetic typing of DR3 and DR4 antigens was performed for all 99 subjects. The diagnosis of CAN was based on the results of a battery of autonomic tests (heart rate at rest, heart rate response to deep breathing, standing up, systolic blood pressure response to standing up and diastolic blood pressure response to isometric exercise). The diagnosis of CAN was confirmed when 2 or more pathological tests were present. Standardised questionnaires were completed to obtain information on demographic data, clinical events, medications and life style.
The study protocol was approved by the local Ethics committee of Lithuanian University of Health Sciences (Kaunas city); all participants provided written informed consent.
Methods of genotyping
DNA was extracted from peripheral blood leukocytes by salt precipitation. The isolated DNA was dissolved in sterile 1xTE buffer. Isolated DNA concentration was measured with a spectrophotometer, and purity was assessed by measuring the optical density.
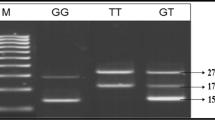
Investigated DNA fragment was amplified with PCR reaction using 100 ng of subjects DNA, dNTP mix, Taq polymerase, 1.25 Dream U and reaction buffer. Specific primers were used for amplification of 213 bp (base pairs) PCR fragment size, which create an artificial restriction fragment length polymorphism in DRB1 gene - DR3 RFLP: 5′ CCG CTG CAC TGT GAA GCT CTC CAC AAC CCC GTA GTT GTG TCT GCA CTAG 3′ and DR4.04: 5′ CGG GTG CGG TTC CTG GAC AGA TAC TTC GAT 3′. PCR was performed using gradient Eppendorf firm thermal cycler. PCR reaction products were visualised on 1.5 % agarose gel. Restriction reaction was performed using 5 units of restriction endonuclease Spe 1 (Enzyme) and 1 Dde (Enzyme) in 15 μl of reaction mixture (composition - 5 μl of the PCR product, restriction endonuclease Spe 1 and 0.5 μl Dde, 1.5 ul 10× reaction buffer and 7.5 μl of water). After 2 h Sau 3A1 enzyme, which recognizes GATC sequence in DR4 allele was added and a fragment of 186 bp is obtained.
Restriction of ACTAGT sequence, which presents only in DR3 or DRB7 allele, was recognized by restriction of endonuclease Spe 1 and obtained fragment of 164 bp. Dde 1 allows to distinguish between DR3 or DRB7 alleles because it recognizes two places of CTNAG in DRB7 allele (codons 51–52 and 58–59), and neither one place in DR3 allele. If it is DRB7 allele Dde 1 cuts PCR into three fragments of 106, 86, and 21 base pairs. If Dde cutting site is located on another allele, there is no the second cutting site, and are presented 106/107 bases duplets.
Restriction fragment size was evaluated in 4 % agarose gel after electrophoresis and colouring EtBr. Gel was documented and restriction fragment sizes were assessed using BioRad gel documentation system.
The assessment of HLA genotyping results:
-
N/N – DRB1 gene does not express neither DR3 nor DR4 antigen (both alleles determine the absence of DR3 and DR4 antigen - homozygote);
-
DR3/DR3 – subject has DR3 antigen (both alleles determine DR3 antigen- homozygote);
-
DR4/DR4 – subject has DR4 antigen (both alleles determine DR4 antigen- homozygous);
-
DR3/DR4 – subject has DR3 and DR4 antigens (heterozygote);
-
DR3/N – subject has DR3 antigen (heterozygote);
-
DR4/N – subject has DR4 antigen (heterozygote).
Statistical analysis
Statistical analysis was performed using SPSS v. 20.0 (License No. 9582494). The sample size was calculated during a pilot study using PASS (Power analysis and sample size software). The power of the study was selected to be β = 0.8, and the confidence level - α = 0.05. Data are presented in absolute value and frequency. Data are presented as means and standard deviations. Used statistical methods: variables of two independent samples were compared using a Mann–Whitney test. Chi-square (χ2) was used for rates comparison. Risk alleles search was performed using univariate odds ratio analysis. Results were interpreted as statistically significant at an error probability of p <0.05.
Results
The 99 patients with T1D (47 males and 52 females) were included in the study. Genotypes in two homogeneous groups according to the age of T1D diagnosis, duration of diabetes, gender and HbA1c were analysed. The characteristics of patients are summarized by AER status in Table 1. The proportion of CAN was significantly greater in case group (6.81 %) as compared with control group (2.32 %) (p = 0.03).
The highest 24 h AER was found in patients with DRB1 gene expressed DR3 risk alleles group. The lowest AER was found in patients with DRB1 gene with no expression of both DR3 and DR4 antigen (Table 2). Assessing DRB1 gene’s N/N, DR3 and DR4 risk alleles combinations and microalbuminuria, the greatest significance was achieved between N/N and DR3 alleles groups.
Genotyping of 99 patients with T1D was performed: no DR3 and DR4 risk alleles were found in 22 (22.22 %) patients, DR3 alleles were present in 47 (47.48 %), DR4 alleles in 25 (25.25 %), and DR3/DR4 alleles in 5 (5.05 %) (Table 3). According to the frequency of different combinations of alleles we didn’t find statistical difference among case (AER 30–300 mg/24 h) and control (AER <30 mg/24 h) groups.
We compared by DRB1 gene’s N/N, DR3 and DR4 risk alleles in patients with T1D, most case patients had DR3 risk alleles: 31 (51.67 %) patient had DR3 alleles, 13 (21.67 %) patients had DR4 alleles and 4 (6.67 %) patients had DR3/DR4 alleles. We confirmed the 1.87 (p = 0.021) increased relative risk for microalbuminuria in patients with DR3/DR3 alleles and the same duration of diabetes (Table 4).
The distribution of DR3 and DR4 risk alleles was not associated with CAN both in patients with normal AER and microalbuminuria (1.6 vs 2.1; p = 0.21).
Discussion
Microalbuminuria is an early sign of diabetic nephropathy. Microalbuminuria is seldom found in patients within first 5 years from onset of T1D, usually microalbuminuria starts at 5 to 15 years of duration of diabetes and then increases over time [6, 7]. In a systematic review of nine longitudinal studies examining moderately increased albuminuria in 7938 patients with T1D, the overall prevalence of moderately increased albuminuria was 28 % at a mean duration of T1D of 15 years [14].
J.H. Warram and co-authors state that microalbuminuria can be found in about 20 % of patients with type 1 diabetes with duration of disease 20 years and in a 50 % with duration of disease more than 30 years [7]. For kids and teens advanced stage of diabetic nephropathy is seldom found [15]. Several studies describe the rapid clinical and histological development of diabetic nephropathy in children and adolescents at 4–11 years duration of disease [16, 17]. Familial tendency to develop this diabetes complication was determined [18], showing the importance of genetic factors. But still no individual gene that results in kidney damage in patients with type 1 diabetes was identified.
The impact of apolipoprotein E gene [19], growth arrest-specific 6 gene (Gas6) and their receptors Ax1 [20], polymorphism of the enzyme V16A of manganese superoxide dismutase [21], insulin gene [22], DQA1 and DQB1 haplotypes [23], HLA-DR3 and DR4 alleles [24] are investigated to clarify the reasons for the development of diabetic nephropathy. HLA has a significant impact not only to the development of T1D, but also to diabetes complications. The aim of the study was to evaluate a link between HLA-DR3/DR4 and microalbuminuria among patients with the same durations of diabetes.
The highest risk DR/DQ haplotypes for T1D are DR3-DQA1*0501-DQB1*0201 (DR3) and DR4-DQA1*0301-DQB1*0302 (DR4), and these alleles account for 30–50 % of genetic T1D risk [10]. Although 40 % of whites in the US have an HLA-DR3 or -DR4 allele, at least 1 of these alleles is present in 95 % of patients with T1D. The estimated risk of developing T1D for the general population in children who have the HLA-DR3/4 genotype is approximately 1 in 15 to 1 in 25 vs. a risk of 1 in 300 in the general population [25]. Only 2.4 % of the general population carries this genotype compared to 30–40 % of T1D patients. Although this genotype confers extremely high risk, there is a spectrum of risk associated with HLA DR/DQ genotypes—from increased, to neutral, to protective [26].
Very early small studies looked at HLA in people with diabetic nephropathy, but no reliable associations was found [27–29]. The assumption that the HLA DRB1*04 alleles may protect against development of diabetic nephropathy appeared recently. A. Svejgaard et al. investigated 71 patients with long duration of T1D, 24 had diabetic nephropathy and 51 normal AER, and found a marked protective effect of DR4 allele (p = 0.02) [30]. In our study we have found the highest risk of increased AER in T1D patients with DRB1 gene expressed DR3 risk allele. Also the lowest AER we found in patients with neither DR3 nor DR4 antigen expressed. Genetics of Kidneys in Diabetes (GoKinD) study [31], examined patients with T1D with nephropathy (n = 829) and without nephropathy (n = 904). Results of this study showed, that diabetic probands who were homozygous for HLA DRB1*04 were 50 % less likely to have nephropathy than probands without any DRB1*04 alleles. In heterozygous carriers, a protective effect of this allele was not as clearly evident. This association was seen in probands with both short (<28 years, p = 0.02) and long (≥28 years, p = 0.0001) duration of diabetes. Interestingly, DRB1*04 appears to be both a risk allele for T1D and a protective allele for nephropathy. In our study patients who were homozygous for HLA DRB1*04 were 50 % less likely to have microalbuminuria than patients who were homozygous for HLA DRB1*03 allele. The main weakness of our study is the small sample size. The power of the study was selected to be β = 0.8, and the confidence level - α = 0.05. We calculated that we need to find 334 patients (case and controls). Finally we could include and analyze only 99 patients, so further research is necessary to confirm our results. The other limitation is that a population of our study was only caucasians, and possibly our findings couldn’t be applied to other races.
Conclusions
The 1.87 (p = 0.021) increased relative risk for microalbuminuria was found in patients with DR3/DR3 alleles and the same duration of diabetes.
Availability of data and materials
Data supporting our findings can be found at http://www.lsmuni.lt/en/library/about-library-and-information-centre/, http://www.kmu.lt/endokrinologija/index.php?psl=pradzia and http://vddb.library.lt/obj/LT-eLABa-0001:E.02~2011~D_20110922_122348-00650.
Abbreviations
- AER:
-
albumin excretion rate
- CAN:
-
cardiovascular autonomic neuropathy
- HLA:
-
human leukocyte antigen
- MHC:
-
the Major Histocompatibility Complex
- T1D:
-
type 1 diabetes
References
Brorsson C, Hansen NT, Lage K, Bergholdt R, Brunak S, Pociot F. Identification of T1D susceptibility genes within the MHC region by combining protein interaction networks and SNP genotyping data. Diabetes Obes Metab. 2009;11 Suppl 1:60–6.
Kazanavicius G, Barsiene L, Cibas P, Danytė E, Daugintyte-Petrosiene L, Graziene A. Endocrinology. 2009:90–97; 106–107; 167–174.
Groop PH, Thomas MC, Moran JL, Waden J, Thorn LM, Makinen VP, Rosengard-Barlund M, Saraheimo M, Hietala K, Heikkila O, et al. The presence and severity of chronic kidney disease predicts all-cause mortality in type 1 diabetes. Diabetes. 2009;58(7):1651–8.
Maser RS, Lenhard MJ, DeCherney GS. Cardiovascular autonomic neuropathy: the clinical significance of its determination. Endocrinologist. 2000;10:27–33.
Vergouwe Y, Soedamah-Muthu SS, Zgibor J, Chaturvedi N, Forsblom C, Snell-Bergeon JK, Maahs DM, Groop PH, Rewers M, Orchard TJ, et al. Progression to microalbuminuria in type 1 diabetes: development and validation of a prediction rule. Diabetologia. 2010;53(2):254–62.
Hovind P, Tarnow L, Rossing P, Jensen BR, Graae M, Torp I, Binder C, Parving HH. Predictors for the development of microalbuminuria and macroalbuminuria in patients with type 1 diabetes: inception cohort study. BMJ. 2004;328(7448):1105.
Warram JH, Gearin G, Laffel L, Krolewski AS. Effect of duration of type I diabetes on the prevalence of stages of diabetic nephropathy defined by urinary albumin/creatinine ratio. J Am Soc Nephrol. 1996;7(6):930–7.
Predictors of the development of microalbuminuria in patients with Type 1 diabetes mellitus: a seven-year prospective study. The Microalbuminuria Collaborative Study Group. Diabet Med. 1999;16(11):918–25.
Correa-Giannella ML, Vieira SM. [Genetic susceptibility to microangiopathy development in Type 1 diabetes mellitus]. Arq Bras Endocrinol Metabol. 2008;52(2):375–86.
Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet. 1996;59(5):1134–48.
Steck AK, Rewers MJ. Genetics of type 1 diabetes. Clin Chem. 2011;57(2):176–85.
WHO Expert Committe on Diabetes Mellitus. Second report. Technical Report series no 1985, 742. Geneva: WHO. 1980;646:1–80.
Mogensen CE, Hansen KW, Nielsen S, Pedersen MM, Rehling M, Schmitz A. Monitoring diabetic nephropathy: glomerular filtration rate and abnormal albuminuria in diabetic renal disease–reproducibility, progression, and efficacy of antihypertensive intervention. Am J Kidney Dis. 1993;22(1):174–87.
Newman DJ, Mattock MB, Dawnay AB, Kerry S, McGuire A, Yaqoob M, Hitman GA, Hawke C. Systematic review on urine albumin testing for early detection of diabetic complications. Health Technol Assess. 2005;9(30):iii-vi, xiii-163.
Bogdanovic R. Diabetic nephropathy in children and adolescents. Pediatr Nephrol. 2008;23(4):507–25.
Chobanian MC, Chevalier RL, Sturgill BC, Bolton WK. Early onset of clinical diabetic nephropathy in children–a new subgroup? Int J Pediatr Nephrol. 1984;5(1):23–9.
Francis J, Rose SJ, Raafat F, Milford DV. Early onset of diabetic nephropathy. Arch Dis Child. 1997;77(6):524–5.
Rossing P. Prediction, progression and prevention of diabetic nephropathy. The Minkowski Lecture 2005. Diabetologia. 2006;49(1):11–9.
Li Y, Tang K, Zhang Z, Zhang M, Zeng Z, He Z, He L, Wan C. Genetic diversity of the apolipoprotein E gene and diabetic nephropathy: a meta-analysis. Mol Biol Rep. 2011;38(5):3243–52.
Chandy A, Pawar B, John M, Isaac R. Association between diabetic nephropathy and other diabetic microvascular and macrovascular complications. Saudi J Kidney Dis Transpl. 2008;19(6):924–8.
Mollsten A, Marklund SL, Wessman M, Svensson M, Forsblom C, Parkkonen M, Brismar K, Groop PH, Dahlquist G. A functional polymorphism in the manganese superoxide dismutase gene and diabetic nephropathy. Diabetes. 2007;56(1):265–9.
Bain SC, Chowdhury TA. Genetics of diabetic nephropathy and microalbuminuria. J R Soc Med. 2000;93(2):62–6.
Agardh E, Gaur LK, Lernmark A, Agardh CD. HLA-DRB1, -DQA1, and -DQB1 subtypes or ACE gene polymorphisms do not seem to be risk markers for severe retinopathy in younger Type 1 diabetic patients. J Diabetes Complications. 2004;18(1):32–6.
Saner B, Bonney E, Barbosa J. HLA-DR antigens and diabetic nephropathy. Diabetologia. 2004;25(5):452.
Rewers M, Bugawan TL, Norris JM, Blair A, Beaty B, Hoffman M, McDuffie RS, Jr., Hamman RF, Klingensmith G, Eisenbarth GS et, al. Newborn screening for HLA markers associated with IDDM: diabetes autoimmunity study in the young (DAISY). Diabetologia. 1996;39(7):807–12.
Sheild JPH, Wadsworth EJK, Baum JD. The genetic contribution to disease pathogenesis inchildhood diabetes is greatest in the very young. Diabet Med. 1995;12:377-79.
Barbosa J, Saner B. Do genetic factors play a role in the pathogenesis of diabetic microangiopathy? Diabetologia. 1984;27(5):487–92.
Chowdhury TA, Dyer PH, Mijovic CH, Dunger DB, Barnett AH, Bain SC. Human leucocyte antigen and insulin gene regions and nephropathy in type I diabetes. Diabetologia. 1999;42(8):1017–20.
Ronningen KS, Bangstad HJ, Undlien DE, Thorsby E. Influence of genetic factors (HLA class II genes, insulin-gene region polymorphisms) and metabolic control on the development of diabetic nephropathy. Diabetes Res. 1993;23(1):31–40.
Svejgaard A, Jakobsen BK, Platz P, Ryder LP, Nerup J, Christy M, Borch-Johnsen K, Parving HH, Deckert T, Molsted-Pedersen L, et al. HLA associations in insulin-dependent diabetes: search for heterogeneity in different groups of patients from a homogeneous population. Tissue Antigens. 1986;28(4):237–44.
Cordovado SK, Zhao Y, Warram JH, Gong H, Anderson KL, Hendrix MM, Hancock LN, Cleary PA, Mueller PW. Nephropathy in type 1 diabetes is diminished in carriers of HLA-DRB1*04: the genetics of kidneys in diabetes (GoKinD) study. Diabetes. 2008;57(2):518–22.
Acknowledgements
We thank all the patients participating in our study. We are also grateful to geneticists who performed genetic testing and to the staff of Institute of Endocrinology, Lithuanian University of Health Sciences, who undertook various assessments during the study. This work was supported by Institute of Endocrinology, Lithuanian University of Health Sciences.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DRV contributed to data analysis and wrote manuscript. ED participated in the design of the study and coordination and edited manuscript. RZ conceived of the study, participated in its design, performed statistical analysis and edited manuscript. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ražanskaitė-Virbickienė, D., Danytė, E. & Žalinkevičius, R. HLA-DRB1*03 as a risk factor for microalbuminuria in same duration of type 1 diabetes: a case control study. BMC Nephrol 17, 38 (2016). https://doi.org/10.1186/s12882-016-0252-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12882-016-0252-4