Abstract
Background
Pathogenic variants associated with hereditary breast cancer have been reported for BRCA1 and BRCA2 (BRCA1/2) genes in patients from multiple ethnicities, but limited information is available from sub-Saharan African populations. We report a BRCA2 pathogenic variant in a Senegalese family with hereditary breast cancer.
Methods
An index case from a consanguineous family and nineteen healthy female relatives were recruited after informed consent. Along with this family, 14 other index cases with family history of breast cancer were also recruited. For the control populations we recruited 48 healthy women with no cancer diagnosis and 48 women diagnosed with sporadic breast cancer without family history. Genomic DNA was extracted from peripheral blood. All BRCA2 exons were amplified by PCR and sequenced. Sequences were compared to the BRCA2 GenBank reference sequence (NM_000059.3) using Alamut Software.
Results
We identified a novel nonsense pathogenic variant c.5219 T > G; p.(Leu1740Ter) in exon 11 of BRCA2 in the index case. The pathogenic variant was also identified in three sisters and one daughter, but was absent in the controls and unrelated cases.
Conclusions
This is the first report of a novel BRCA2 pathogenic variant in a Senegalese family with hereditary breast cancer. This result confirms the diversity of hereditary breast cancer pathogenic variants across populations and extends our knowledge of genetic susceptibility to breast cancer in Africa.
Similar content being viewed by others
Background
Breast cancer is the leading female cancer in the world in terms of incidence and mortality. In 2012 it was diagnosed in 1.7 million cases with 500,000 deaths [1]. Its standardized incidence is estimated at 36.1 per 100,000 women in West Africa [2]. In Senegal, as in most Sub-Saharan African (SSA) countries, breast cancer is the second most common cancer in women after cervical cancer, and may now be the most common cancer based on preliminary reports [2]. It is currently estimated that 5–10% of breast cancers are due to an inherited predisposition [3,4,5]. Approximately 20–25% of this risk is associated to pathogenic variants of two high penetrance susceptibility genes, BRCA1 and BRCA2, located on chromosomes 17q21 and 13q12, respectively [6, 7]. Both genes are involved in DNA repair and other important biological functions [8]. Risk increases with the number of affected women within the family, early age at diagnosis and the degree of relationship with other affected women [9,10,11]. The cumulative risk of breast cancer by age 80 years was estimated to 72% for BRCA1 carriers and 69% for BRCA2 carriers. For ovarian cancer the cumulative risk by age 80 years was estimated to 44% for BRCA1 carriers and 17% for BRCA2 carriers [10,11,12,13]. The human BRCA2 gene contains 27 exons, among which exon 11 is the largest. The coding sequence (Refseq transcript mRNA: NM_000059.3) size is 11,386 bp and it encodes a protein of 3418 amino acids (Refseq protein NP_000050) [14]. Genetic variation analysis of BRCA2 has identified a large number of different pathogenic germline variants in breast cancer patients and more than a thousand different disease–causing germline pathogenic variants were listed in the Breast Cancer Information Core Database (BIC; http://research.nhgri.nih.gov/bic/) and in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/). Most BRCA2 pathogenic variants have been reported in individuals of European and Asian origin while limited information is available on SSA populations [15,16,17,18,19]. Herein we report a novel pathogenic variant in BRCA2 in a consanguineous family with a family history of breast cancer.
Methods
Study population
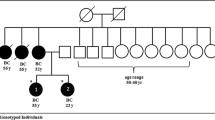
A female index case from a family with a consanguineous mating (Fig. 1) and 19 healthy relative women were recruited after informed consent. The index case was a 53-years-old woman of Wolof ethnicity who died at age 54 from moderately differentiated triple negative ductal adenocarcinoma of the right breast, clinical stage T4dN1M0, SBRII grade. Therapeutic management at the Joliot Curie Institute of Hospital Le Dantec in Dakar, Senegal, consisted of 7 courses of chemotherapy followed by surgical removal of the right breast and radiotherapy. She died as a result of a brain metastasis. Her mother sister, mother’s cousin and maternal grandmother’s sister, also died from breast cancer. The index case was married to her mother’s cousin (Fig. 1). Along with this family, 14 other unrelated index cases with family history of breast cancer, were also recruited. For the control populations we recruited 48 healthy women with no cancer diagnosis (mean age 40.7 years) who came for routine checkup at the Laboratory of Biology of Le Dantec Hospital, and 48 women diagnosed with sporadic breast cancer without family history (mean age at diagnosis 45.7 years) from the Joliot Curie Institute of Le Dantec Hospital. This study was approved by the ethics committee of Cheikh Anta DIOP University under Protocol 014/2014 / CER / UCAD.
BRCA1 and BRCA2 screening
For each participant a 5 ml blood sample was collected in EDTA tubes. Genomic DNA was extracted from whole blood with a QIAamp® DNA blood Mini kit (Qiagen, Hildel, Germany). In the proband from the consanguineous family as well as in the remaining 14 index cases with familial breast cancer, BRCA1 and BRCA2 exons were amplified by PCR with specific primers located in the introns, flanking the intron/exon boundaries. Twenty-eight fragments covering the 22 coding exons of BRCA1 and 32 fragments covering the 26 coding exons of BRCA2 gene were amplified [20]. The large exons 10 and 11 of BRCA2 were amplified as 2 and 9 fragments respectively while exon 11 of BRCA1 was amplified as 7 fragments (see Additional file 1: Tables S1 and S2). PCRs were carried out with initial denaturation at 95 °C for 10 min followed by 40 cycles of 95 °C for 30s, 55 °C for 30s, and 72 °C for 30s with a GeneAmp® PCR System 9700 (Applied Biosystems) as described previously [20]. The PCR products were purified with a MinElute 96UF kit and sequenced using a Big Dye terminator V3.1 sequencing kit on a 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Both forward and reverse strands were sequenced. The sequences were compared to the BRCA2 GenBank reference sequence (NM_000059.3) with Alamut Software. For the control populations (healthy and sporadic breast cancer groups) and healthy relatives of the proband, only the targeted fragment containing the pathogenic BRCA2 variant identified in the proband, was sequenced. DNAs from three healthy relatives were of poor quality and were not sequenced.
Results
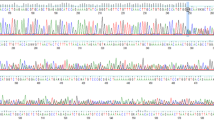
No pathogenic variants were identified in BRCA1 in the proband. A BRCA2 pathogenic variant, namely c.5219 T > G; p.(Leu1740Ter) (according to the HGVS nomenclature), was identified in the proband. This variant is located in exon 11 of the gene and is predicted to introduce a premature stop codon at position 1740 of the BRCA2 protein. The chromatogram is shown in (Fig. 2).
This pathogenic variant was also detected in three sisters and one daughter of the index case (Fig. 1). The variant was likely transmitted to the index case by her mother (III-2) even though she was not diagnosed with breast cancer, but has a sister who died of breast cancer. The index case’s mother in turn would likely have inherited this pathogenic variant from the grandmother (II-1) who’s sister died of breast cancer (II-3). Four healthy relatives of the index case had the pathogenic variant and would be at risk of developing breast cancer.
This pathogenic variant was not detected in any participant from the two control groups (healthy and sporadic breast cancer groups). This variant has not been described previously in BIC and ClinVar databases, nor in the literature. Other single nucleotide variants were identified in the index case in different BRCA2 exons (Table 1). These variants have been described in the literature and are classified as benign by the expert panel in the ClinVar database and therefore are not pathogenic.
For the other 14 index cases recruited for BRCA1/BRCA2 genetic testing, we identified a recurrent pathogenic variant of the BRCA1 gene in 6 families out of the 14. No pathogenic variant was detected for the remaining 8 families (data not shown).
Discussion
Breast cancer is the most commonly diagnosed type of cancer in women in the world [2]. The epidemiology in SSA countries is characterized by younger age at diagnosis, triple negative histopathology, advanced clinical stage and poor prognosis [21,22,23]. The phenotype of the breast cancer diagnosed in the studied index case matched with this epidemiology. Inherited breast cancer risk is associated to pathogenic variants of two high penetrance susceptibility genes, BRCA1 and BRCA2, yet pathogenic variants in other genes including PALB2, TP53 and PTEN have also been linked with high risk of breast cancer [24, 25]. Pathogenic variants of these genes have been associated with susceptibility to hereditary breast cancer in populations of European and Asian origin [16,17,18, 26,27,28,29] while in Africa, most of the available data come from studies conducted among North African, Nigerian, Sudan and South African populations [16, 30,31,32,33,34,35,36,37,38,39,40].
Few pathogenic variants of BRCA2 gene have been reported in SSA populations. The BRCA2 pathogenic variants identified in African populations and reported in the literature are summarized in Table 2. Most causal pathogenic variants have been identified in exon 11 [16, 31, 33, 35,36,37], and are predominantly deletions or duplications. Only a few nucleotide substitutions leading to premature stop codons have been reported. The novel pathogenic variant we identified in this study is a substitution leading to premature stop codon and is located in exon 11 at position 5219.
Although other pathogenic variants surrounding this position of the BRCA2 gene have been reported in the ClinVar database, any pathogenic variant involving the particular codon has been reported in SSA populations (Table 3). These pathogenic variants lead to stop codons or frameshift at amino acid 1739, 1740 or 1741 of BRCA2 protein.
Several studies have shown that the pathogenic variant spectrum identified in black populations is different from Caucasian populations [19, 29, 47, 48]. The c.2808_2811delACAA pathogenic variant in BRCA2 was frequently reported in European populations [49], and was reported only once in a young black girl with breast cancer in Ibadan, Nigeria [27]. The pathogenic variant we identified in BRCA2 was detected in one family out of 15 recruited for BRCA1/2 genetic testing, while we identified a recurrent pathogenic variant of the BRCA1 gene in 6 families out of the 15 (data not shown). These observations confirm the diversity of pathogenic variants between populations but also within the same population [19, 37, 50].
It has also been reported that Fanconi Anemia (FA) is caused by biallelic FANCD1/BRCA2 pathogenic variants [51]. In this family it was unclear whether there was family member with Fanconi anemia like symptoms or early death, or not.
Conclusions
We report a novel pathogenic variant c.5219 T > G p.(Leu1740Ter) in BRCA2 in a consanguineous Senegalese family with a family history of breast cancer. This result highlights the diversity of hereditary breast cancer pathogenic variants and extends the knowledge of genetic susceptibility to breast cancer in Africa. The benefits of clinical genetic testing of BRCA1/2 in prevention and personalised treatment is unquestionable and it should be adapted to each population’s intrinsic genetic characteristics.
Abbreviations
- BIC:
-
Breast Cancer Information Core Database
- BRCA1/2 :
-
Breast cancer gene 1 / 2
- DNA:
-
Desoxyribonucleic Acid
- EDTA:
-
Ethylene diamine tetra acetic acid
- G:
-
Guanine
- HGVS:
-
Human Genome Variation Society
- NCBI:
-
National Center for Biotechnology Information
- PCR:
-
Polymerase Chain Reaction
- RefSeq:
-
Reference Sequence
- SBR:
-
Scarff-Bloom-Richardson
- SNP:
-
Single nucleotide polymorphism
- SSA:
-
Sub Saharan Africa
- T:
-
Thymine
- Ter:
-
Termination stop codon
References
Stuart J, Schnitt SRL. World Cancer Report. In: IARC Library Cataloguing in Publication Data; 2014. p. 362–72.
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86.
Fackenthal JD, Sveen L, Gao Q, Kohlmeir EK, Adebamowo C, Ogundiran TO, Adenipekun AA, Oyesegun R, Campbell O, Rotimi C, et al. Complete allelic analysis of BRCA1 and BRCA2 variants in young Nigerian breast cancer patients. J Med Genet. 2005;42(3):276–81.
Valeria Viassolo AA, Chappuisa PO. Breast cancer: genetic risk. Imagerie de la Femme. 2016;26:15.
Golubnitschaja O, Debald M, Yeghiazaryan K, Kuhn W, Pesta M, Costigliola V, Grech G. Breast cancer epidemic in the early twenty-first century: evaluation of risk factors, cumulative questionnaires and recommendations for preventive measures. Tumour Biol. 2016;37(10):12941–57.
Tonin P, Serova O, Simard J, Lenoir G, Feunteun J, Morgan K, Lynch H, Narod S. The gene for hereditary breast-ovarian cancer, BRCA1, maps distal to EDH17B2 in chromosome region 17q12-q21. Human molecular genetics. 1994;3(9):1679–82.
Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, Nguyen K, Seal S, Tran T, Averill D, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265(5181):2088–90.
Fradet-Turcotte A, Sitz J, Grapton D, Orthwein A. BRCA2 functions: from DNA repair to replication fork stabilization. Endocr Relat Cancer. 2016;23(10):T1–T17.
Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–30.
Antoniou AC, Cunningham AP, Peto J, Evans DG, Lalloo F, Narod SA, Risch HA, Eyfjord JE, Hopper JL, Southey MC, et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer. 2008;98(8):1457–66.
Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25(11):1329–33.
Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, Evans DG, Izatt L, Eeles RA, Adlard J, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst. 2013;105(11):812–22.
Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, Jervis S, van Leeuwen FE, Milne RL, Andrieu N, et al. Risks of breast, ovarian, and contralateral breast Cancer for BRCA1 and BRCA2 mutation carriers. Jama. 2017;317(23):2402–16.
Easton DF, Bishop DT, Ford D, Crockford GP. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. The breast Cancer linkage consortium. Am J Hum Genet. 1993;52(4):678–701.
Anim JT. Breast cancer in sub-Saharan African women. Afr J Med. 1993;22(1):5–10.
Francies FZ, Wainstein T, De Leeneer K, Cairns A, Murdoch M, Nietz S, Cubasch H, Poppe B, Van Maerken T, Crombez B, et al. BRCA1, BRCA2 and PALB2 mutations and CHEK2 c.1100delC in different south African ethnic groups diagnosed with premenopausal and/or triple negative breast cancer. BMC Cancer. 2015;15:912.
Oluwagbemiga LA, Oluwole A, Kayode AA. Seventeen years after BRCA1: what is the BRCA mutation status of the breast cancer patients in Africa? - a systematic review. SpringerPlus. 2012;1(1):83.
Ossa CA, Torres D. Founder and recurrent mutations in BRCA1 and BRCA2 genes in Latin American countries: state of the art and literature review. Oncologist. 2016;21(7):832–9.
Rebbeck TR, Friebel TM, Friedman E, Hamann U, Huo D, Kwong A, Olah E, Olopade OI, Solano AR, Teo SH, et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum Mutat. 2018;39:593–620.
Noguchi T, Bourdon V, Sobol H. About sequence quality: impact on clinical applications. Genetic testing and molecular biomarkers. 2014;18(5):299–305.
Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, et al. Race, breast cancer subtypes, and survival in the Carolina breast Cancer study. Jama. 2006;295(21):2492–502.
Kantelhardt EJ, Muluken G, Sefonias G, Wondimu A, Gebert HC, Unverzagt S, Addissie A. A review on breast Cancer care in Africa. Breast Care (Basel). 2015;10(6):364–70.
Ly M, Antoine M, Andre F, Callard P, Bernaudin JF, Diallo DA. Breast cancer in Sub-Saharan African women: review. Bulletin du cancer. 2011;98(7):797–806.
Antoniou AC, Foulkes WD, Tischkowitz M. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371(17):1651–2.
Apostolou P, Fostira F. Hereditary breast cancer: the era of new susceptibility genes. Biomed Res Int. 2013;2013:747318.
Gao Q, Tomlinson G, Das S, Cummings S, Sveen L, Fackenthal J, Schumm P, Olopade OI. Prevalence of BRCA1 and BRCA2 mutations among clinic-based African American families with breast cancer. Hum Genet. 2000;107(2):186–91.
Gao Q, Adebamowo CA, Fackenthal J, Das S, Sveen L, Falusi AG, Olopade OI. Protein truncating BRCA1 and BRCA2 mutations in African women with pre-menopausal breast cancer. Hum Genet. 2000;107(2):192–4.
Nanda R, Schumm LP, Cummings S, Fackenthal JD, Sveen L, Ademuyiwa F, Cobleigh M, Esserman L, Lindor NM, Neuhausen SL, et al. Genetic testing in an ethnically diverse cohort of high-risk women: a comparative analysis of BRCA1 and BRCA2 mutations in American families of European and African ancestry. JAMA. 2005;294(15):1925–33.
Haffty BG, Choi DH, Goyal S, Silber A, Ranieri K, Matloff E, Lee MH, Nissenblatt M, Toppmeyer D, Moran MS. Breast cancer in young women (YBC): prevalence of BRCA1/2 mutations and risk of secondary malignancies across diverse racial groups. Ann Oncol. 2009;20(10):1653–9.
Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer. 2007;7(12):937–48.
Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat. 2011;125(2):325–49.
van der Merwe NC, Hamel N, Schneider SR, Apffelstaedt JP, Wijnen JT, Foulkes WD. A founder BRCA2 mutation in non-Afrikaner breast cancer patients of the Western cape of South Africa. Clin Genet. 2012;81(2):179–84.
Seymour HJ, Wainstein T, Macaulay S, Haw T, Krause A. Breast cancer in high-risk Afrikaner families: is BRCA founder mutation testing sufficient? S Afr Med J. 2016;106(3):264–7.
Schoeman M, Apffelstaedt JP, Baatjes K, Urban M. Implementation of a breast cancer genetic service in South Africa - lessons learned. S Afr Med J. 2013;103(8):529–33.
Henouda S, Bensalem A, Reggad R, Serrar N, Rouabah L, Pujol P. Contribution of BRCA1 and BRCA2 germline mutations to early Algerian breast Cancer. Dis Markers. 2016;2016:7869095.
Awadelkarim KD, Aceto G, Veschi S, Elhaj A, Morgano A, Mohamedani AA, Eltayeb EA, Abuidris D, Di Gioacchino M, Battista P, et al. BRCA1 and BRCA2 status in a central Sudanese series of breast cancer patients: interactions with genetic, ethnic and reproductive factors. Breast Cancer Res Treat. 2007;102(2):189–99.
Fackenthal JD, Zhang J, Zhang B, Zheng Y, Hagos F, Burrill DR, Niu Q, Huo D, Sveen WE, Ogundiran T, et al. High prevalence of BRCA1 and BRCA2 mutations in unselected Nigerian breast cancer patients. Int J Cancer. 2012;131(5):1114–23.
Bensam M, Hafez E, Awad D, El-Saadani M, Balbaa M. Detection of new point mutations of BRCA1 and BRCA2 in breast cancer patients. Biochem Genet. 2014;52(1–2):15–28.
Tazzite A, Jouhadi H, Nadifi S, Aretini P, Falaschi E, Collavoli A, Benider A, Caligo MA. BRCA1 and BRCA2 germline mutations in Moroccan breast/ovarian cancer families: novel mutations and unclassified variants. Gynecol Oncol. 2012;125(3):687–92.
Troudi W, Uhrhammer N, Sibille C, Dahan C, Mahfoudh W, Bouchlaka Souissi C, Jalabert T, Chouchane L, Bignon YJ, Ben Ayed F, et al. Contribution of the BRCA1 and BRCA2 mutations to breast cancer in Tunisia. J Hum Genet. 2007;52(11):915–20.
Fourati A, Louchez MM, Fournier J, Gamoudi A, Rahal K, El May MV, El May A, Revillion F, Peyrat JP. Screening for common mutations in BRCA1 and BRCA2 genes: interest in genetic testing of Tunisian families with breast and/or ovarian cancer. Bull Cancer. 2014;101(11):E36–40.
Zhang J, Fackenthal JD, Zheng Y, Huo D, Hou N, Niu Q, Zvosec C, Ogundiran TO, Hennis AJ, Leske MC, et al. Recurrent BRCA1 and BRCA2 mutations in breast cancer patients of African ancestry. Breast Cancer Res Treat. 2012;134(2):889–94.
Cherbal F, Bakour R, Adane S, Boualga K, Benais-Pont G, Maillet P. BRCA1 and BRCA2 germline mutations screening in Algerian breast/ovarian cancer families. Dis Markers. 2010;28(6):377–84.
Laarabi FZ, Jaouad IC, Ouldim K, Aboussair N, Jalil A, Gueddari BE, Benjaafar N, Sefiani A. Genetic testing and first presymptomatic diagnosis in Moroccan families at high risk for breast/ovarian cancer. Oncol Lett. 2011;2(2):389–93.
Guaoua S, Ratbi I, Lyahyai J, El Alaoui SC, Laarabi FZ, Sefiani A. Novel nonsense mutation of BRCA2 gene in a Moroccan man with familial breast cancer. Afr Health Sci. 2014;14(2):468–71.
Riahi A, Kharrat M, Ghourabi ME, Khomsi F, Gamoudi A, Lariani I, May AE, Rahal K, Chaabouni-Bouhamed H. Mutation spectrum and prevalence of BRCA1 and BRCA2 genes in patients with familial and early-onset breast/ovarian cancer from Tunisia. Clin Genet. 2015;87(2):155–60.
Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266(5182):120–2.
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66–71.
Neuhausen SL, Godwin AK, Gershoni-Baruch R, Schubert E, Garber J, Stoppa-Lyonnet D, Olah E, Csokay B, Serova O, Lalloo F, et al. Haplotype and phenotype analysis of nine recurrent BRCA2 mutations in 111 families: results of an international study. Am J Hum Genet. 1998;62(6):1381–8.
Eley JW, Hill HA, Chen VW, Austin DF, Wesley MN, Muss HB, Greenberg RS, Coates RJ, Correa P, Redmond CK, et al. Racial differences in survival from breast cancer. Results of the National Cancer Institute black/white Cancer survival study. Jama. 1994;272(12):947–54.
Loizidou MA, Hadjisavvas A, Tanteles GA, Spanou-Aristidou E, Kyriacou K, Christophidou-Anastasiadou V. Fanconi anemia-D1 due to homozygosity for the BRCA2 gene Cypriot founder mutation: a case report. Oncol Lett. 2016;11(1):471–3.
Acknowledgements
We thank all family members and controls who agreed to participate in this study. We also thank the staff of the Joliot Curie Institute in Dakar, the Laboratory of Molecular Oncogenetics of the Paoli-Calmette Institute at Marseille, the Laboratory of Cytology, Cytogenetics and Reproductive Biology at Le Dantec Hospital, Dakar, Senegal.
Funding
This work has been funded by the Ministry of Higher Education and Scientific Research of Senegal (FIRST) and the African Center of Excellence for Maternal and Child Health (CEA-SAMEF).
Both funds have contributed to participants recruitment, samples collection, data analysis and interpretation.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Confidential patient data are not shared.
Author information
Authors and Affiliations
Contributions
RND conceived and designed the experiments, JPDD and VBHV performed the experiments and analyzed the data, RND and JPDD wrote the manuscript, AD1- DD- MMD- SK, are oncologists and participated in family recruitment, SAB, YD, BM, AT, OF, PAD, HS and AD2 gave advise in manuscript writing. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the ethics committee of the Cheikh Anta DIOP University under the following number “Protocol 014/2014 / CER / UCAD”. All participants gave their informed written consent before participation in the study. For the deceased patients, the head of the family (Individual IV-15 in the pedigree) gave consent on their behalf.
Consent for publication
All authors have read the manuscript and gave their verbal consent for publication. Each adult participant gave his/her own written consent. For minors and deceased persons, the head of the family (Individual IV-15 in the pedigree) gave consent on their behalf.
Competing interests
The authors declare that they do not have any competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Ndiaye R BRCA2 Supplementary Material. Table S1 Primers used for BRCA1 coding exons PCR amplification. Table S2 Primers used for BRCA2 coding exons PCR amplification. (DOCX 26 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Diop, J.P.D., Diallo, R.N., Bourdon-Huguenin, V. et al. Novel BRCA2 pathogenic variant c.5219 T > G; p.(Leu1740Ter) in a consanguineous Senegalese family with hereditary breast cancer. BMC Med Genet 20, 73 (2019). https://doi.org/10.1186/s12881-019-0814-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-019-0814-y