Abstract
Background
Wiskott-Aldrich syndrome is an X-linked recessive immunodeficiency due to mutations in Wiskott-Aldrich syndrome (WAS) gene. WAS gene is encoded for a multifunctional protein with key roles in actin polymerization, signaling pathways, and cytoskeletal rearrangement. Therefore, the impaired protein or its absence cause phenotypic spectrum of the disease. Since identification of novel mutations in WAS gene can help uncover the exact pathogenesis of Wiskott-Aldrich syndrome, the purpose of this study was to investigate disease causing-mutation in an Iranian male infant suspicious of this disorder.
Case presentation
The patient had persistent thrombocytopenia from birth, sepsis, and recurrent gastrointestinal bleeding suggestive of both Wiskott-Aldrich syndrome and chronic colitis in favor of inflammatory bowel disease (IBD). To find mutated gene in the proband, whole exome sequencing was performed for the patient and its data showed a novel, private, hemizygous splice site mutation in WAS gene (c.360 + 1G > C).
Conclusions
Our study found a novel, splice-site mutation in WAS gene and help consider the genetic counselling more precisely for families with clinical phenotypes of both Wiskott-Aldrich syndrome and inflammatory bowel disease and may suggest linked pathways between these two diseases.
Similar content being viewed by others
Background
Wiskott-Aldrich syndrome (WAS), [MIM: 301000], which is a rare X-linked recessive immunodeficiency is mainly characterized by thrombocytopenia, eczema, infection, and bloody diarrhea. The disease usually leads to death before the age of 10 years [1,2,3]. Patients with WAS, usually, suffer from upper and lower respiratory tract infections, chronic diarrhea, melena and inflammatory bowel disease (IBD). The incidence of WAS throughout the world is estimated to be 1 to 10 in 1 million live birth per year [4]; however; its prevalence or incidence in Iran has not been reported yet. The disease is caused by pathogenic mutations in the WAS gene (located on Xp11.22–23) consisted of 12 exons. The gene is encoded for the Wiskott-Aldrich syndrome protein (WASp), a cytoplasmic 502–amino acid protein involved in the signal transduction from cell surface receptors to the actin cytoskeleton. Its main expression is in non-erythroid hematopoietic cells, indicating its essential roles in the function of these cells [5,6,7]. The WASp is a multifunctional protein with the key involvement in actin polymerization, signaling pathways, and cytoskeletal rearrangement, which is crucial for the monocytes and macrophages migration to infection sites and inflamed tissues, and binding and phagocytosis of antigens [1, 8, 9]. Thus, complete or partial deficiency of WASp leads to malfunctioning of tissue macrophages, neutropenia, and small platelet sizes, causing repeated infections as well as bleeding tendency [4].
Up to know, more than 431 mutations have been reported in WAS gene (Human Genome Mutation Database, HGMD, http://www.hgmd.cf.ac.uk). According to the HGMD database, most of WASp mutations have occurred within the 4 most N-terminal exons of the gene with the arginine residue at position 86 accounting for the most common mutated amino acid in WASP. Different mutations in this protein can cause variable severity of the disease. For instance, study conducted by Greer et al. [10] revealed that the WAS patients with milder clinical presentations had missense mutations. Moreover, study conducted by Villa et al. showed that some WAS mutations can cause only thrombocytopenia with small-sized platelets [2] but, up to now, it is not fully understood why specific disease-causing variants only affect the megakaryocytic cells. In addition, in a WAS family with thrombocytopenia, increased levels of serum IgA and mild nephropathy has been observed to complicate the pathogenesis of this disorder [11]. Since identification of different mutations in WAS gene may help understand the pathogenesis of WAS, the aim of this study was to identify pathogenic variation in our patient with clinical findings suspicious of WAS.
Case presentation
An 8-month-old Iranian male infant, a product of consanguineous marriage, was admitted to our center with history of persistent thrombocytopenia from birth, sepsis, and recurrent gastrointestinal bleeding. In family history, the proband had a sibling who died with similar phenotypes. Initial laboratory findings at different ages were suggestive of idiopathic thrombocytopenic purpura (ITP) (Table 1), therefore; intravenous immunoglobulin (IVIG) was administered for him. At the age of 1 month, he showed mild skin thickening and bone marrow aspiration revealed moderate hypo-cellular marrow with decreased megakaryocyte. However, TORCH study, rheumatologic work up, and levels of complement components such as C3, C4, and CH50 were in normal range. At the age of 4 months, he had increased levels of erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP) and decreased levels of hemoglobin (Hb) and mean platelet volume (MPV), indicating thrombocytopenia. Therefore, IVIG and platelet were administered for the patient.
Based on the patient history and clinical and laboratory findings described above, WAS disease was clinically suspected; therefore, we performed immunological assays. For instance, flow-cytometry showed normal results but the level of antibodies for IgG, IgA, and IgE was high (IgM was in normal range) (Table 2). At that time, to control the sepsis, broad spectrum of antibiotics (Vancomycin and Meropenem) were administered for the patient. At the age of 8 months, the patient had poor feeding, abdominal distension, and lower gastrointestinal bleeding. At that time, the patient was febrile and he was in respiratory distress. Generalized skin petechia and perianal skin tag were also detected. Due to rectal bleeding, endoscopy and sigmoidoscopy were also performed and results revealed severe erythema, erosion, and nodularity in antrum of stomach and nodularity and erythema in the bulb of esophagus. In sigmoidoscopy, skin tag, fistula, fissure in the perianal area, severe and diffused ulcer, and polypoid lesion and decreased vascularity in rectum were detected (Fig. 1a). Colon and rectum biopsy showed chronic colitis with severe activity in favor of inflammatory bowel disease (Fig. 1b). Histological examination of biopsies from the patient also revealed cryptitis and crypt abscesses (Fig. 1c). Since there was no an infectious etiology, these observations were in support of chronic colitis and/or IBD. To find the genetic cause of the disease in our patient, we performed Next Generation Sequencing (NGS) technique to sequence all exons of protein-coding genes.
Whole Exome Sequencing (WES) using Illumina NextSeq500 for pair-end 150-nucleotide sequencing was performed on DNA sample from proband. The NGS results were analyzed using bioinformatics tools, including BWA aligner [12], GATK [13], Annovar [14], and open access bioinformatics tools. Totally, more than 120 K annotated variants were identified with hetero/homo ratio of 1.6 to 1.8, which were then filtered based on their frequency, position, functional consequences, pattern of inheritance and mainly clinical phenotype. NGS data of the proband, revealed a novel, private, hemizygous splice site mutation (NM_000377:exon3:c.360 + 1G > C, Chromosome X) in WAS gene. This identified mutation was not reported previously, therefore, is categorized as the variant of unknown significance (VUS). However, due to lack of any other mutation that can explain the phenotype in this patient, almost complete phenotypic correlation between the disease and identified mutation, and disrupting nature of splice-site mutations, we believe this is a pathogenic mutation. We will deposit into the Clinar database (submission number: SUB3969419). Additionally, as the mutation has no reported frequency in our database (Bayangene) or any other available public variant databases, it was considered as a private mutation.
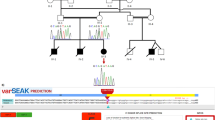
To confirm the identified novel mutation, genomic DNA was prepared from whole blood samples of family members of the proband by QIAamp DNA Blood Mini Kit (Qiagen, Germany) according to the company’s protocol. Then, PCR was carried out for the proband and his parents using following primers: FWAS-E3: GCTCCCAAATCCAGACAC and RWAS-E3: CTTGCACTAGAGGACTCAC (PCR product: 561 bp) for amplification of exon 3 of WAS gene. After that, Sanger sequencing was performed for PCR products on 3130XL Genetic Analyzer (Applied Biosystems USA) according to ABI BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, USA). Sanger sequencing results were analyzed with the use of NCBI BLAST and CodonCode Aligner software in which it confirmed hemizygous status in proband, heterozygous in his mother and normal in his father (Fig. 2). Different bioinformatics software and websites were also used to identify the features and the consequences of mutation in the given position of the protein and also to provide the family pedigree, including Human Splicing Finder (HSF, http://www.umd.be/HSF3/technicaltips.html), STRING (functional protein association networks, https://string-db.org), and Pedigree Chart Designer from CeGat (https://www.cegat.de). Following evidences can confirm that this mutation is led to WAS: 1- WES detected only this mutation to be linked with observed clinical and laboratory findings suggestive of WAS in the proband. 2- Sanger data given in Fig. 2, confirmed the presence of the mutation in the proband as hemizygous, his mother as heterozygous, and his father as normal, confirming the X-linked segregation of the disease. 3- Mutation is located within the donor splice site which is expected to be highly damaging since another mutation in this nucleotide (c.360 + 1G > T, with ID number of CS972885 and CS961711in HGMD) could affect splice site. 4- Using HSF tool, it predicted that the wild type (WT) splice site will be broken and alteration of the WT donor site, most probably affects splicing. 5- Exon 3 is within WH1 domain which is important for binding to a Pro-rich ligand; therefore, skipping of this exon can be very damaging and most probably cause the sever form of the disease [1].
In our study we also used STRING tool to predict the association network between WAS protein and other proteins. Its results revealed that WASp has interaction with several main proteins involved in different biological pathways and cellular component.
Discussion and conclusion
WASp is a member of WAS family of proteins with similar domain named proline-rich region (PPP) which can bind by SH3 domains. WASp has also other essential domains and with identification of various motifs and regions across WASp, it has revealed that they can interact with key proteins in biological pathways. The domains are regulated by different factors such as GTPase, Cdc42 (role in the actin filament formation), ARP2/3 (the cytoskeletal organizing complex), p47nck, a 47-kD cytosolic adapter protein, and tyrosine-protein kinase Fyn [15, 16]. WASp is considered as a link between CDC42 and the actin cytoskeleton since in males affected with WAS, T- lymphocytes show a sever disruption of actin cytoskeleton, which may be due to the impaired Cdc42 signaling [16, 17]. Therefore, dysregulation of pathways linked to WASp have dramatic effects on different biological processes such as actin polymerization [16, 18, 19]. As described in result section, STRING tool showed that WASp has interaction with several key proteins, for example ARP2 actin-related protein 2 homolog, yeast, (ACTR2), WAS/WASL interacting protein family, member 1 (WIPF1), ARP3 actin-related protein 3 homolog, yeast, (ACTR3), actin related protein 2/3 complex, subunit 2, (ARPC2), cell division cycle 42 (GTP binding protein, 25 kDa) (CDC42), actin related protein 2/3 complex, subunit 3 (ARPC3) and so on. These interactions and associations with different proteins suggest the possible indirect involvement of WASp in various pathways and any interrupted interactions may cause life-threating clinical phenotypes. Therefore, the study of different mutations in this gene and functional effects of the mutations help understand the exact mechanism of this gene in different pathways, explaining why different mutations in this gene lead to some specific characterizations of the disease (for example the identified mutation in our study resulted in both WAS and chronic colitis with severe activity in favor of inflammatory bowel disease), and shed light onto the therapeutic approaches for this disease through its corresponding pathways.
In summary, a novel donor splice site mutation was found in WAS gene in our patient who had both WAS and chronic colitis suggestive of inflammatory bowel disease. Such researches may help consider the genetic counselling more precisely for families with clinical phenotypes suggestive of these diseases and help to uncover the biological pathways linked to WAS gene.
Abbreviations
- ACTR2:
-
ARP2 actin-related protein 2 homolog, yeast,
- ACTR3:
-
ARP3 actin-related protein 3 homolog, yeast
- ARPC2:
-
Actin related protein 2/3 complex, subunit 2
- ARPC3:
-
actin related protein 2/3 complex, subunit 3
- CDC42:
-
cell division cycle 42
- CRP:
-
C-Reactive Protein
- ESE:
-
Exonic Splicing Enhancers
- ESR:
-
Erythrocyte Sedimentation Rate
- Hb:
-
Hemoglobin
- HGMD:
-
Human Genome Mutation Database
- HSF:
-
Human Splicing Finder
- IBD:
-
inflammatory bowel disease
- ITP:
-
idiopathic thrombocytopenic purpura
- IVIG:
-
intravenous immunoglobulin
- MPV:
-
mean platelet volume
- NGS:
-
Next Generation Sequencing
- PPP:
-
proline-rich region
- VUS:
-
variants of unknown significance
- WAS:
-
Wiskott-Aldrich syndrome
- WASp:
-
Wiskott-Aldrich syndrome protein
- WES:
-
Whole Exome Sequencing
- WH1:
-
Wasp homology 1
- WIPF1:
-
WAS/WASL interacting protein family, member 1
- WT:
-
Wild Type
- XLN:
-
X-linked neutropenia
- XLT:
-
X-linked thrombocytopenia
References
Massaad MJ, Ramesh N, Geha RS. Wiskott-Aldrich syndrome: a comprehensive review. Ann N Y Acad Sci. 2013;1285:26–43.
Villa A, Notarangelo L, Macchi P, Mantuano E, Cavagni G, Brugnoni D, Strina D, Patrosso MC, Ramenghi U, Sacco MG, et al. X-linked thrombocytopenia and Wiskott-Aldrich syndrome are allelic diseases with mutations in the WASP gene. Nat Genet. 1995;9(4):414–7.
Zhu Q, Zhang M, Blaese RM, Derry JM, Junker A, Francke U, Chen SH, Ochs HD. The Wiskott-Aldrich syndrome and X-linked congenital thrombocytopenia are caused by mutations of the same gene. Blood. 1995;86(10):3797–804.
Guillen-Rocha N, Lopez-Rocha E, Danielian S, Segura-Mendez N, Lopez-Gonzalez L, Lugo-Reyes SO. Wiskott-Aldrich syndrome. A report of a new mutation. Rev Alerg Mex. 2014;61(3):219–23.
Snapper SB, Rosen FS. The Wiskott-Aldrich syndrome protein (WASP): roles in signaling and cytoskeletal organization. Annu Rev Immunol. 1999;17:905–29.
Derry JM, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78(4):635–44.
Binder V, Albert MH, Kabus M, Bertone M, Meindl A, Belohradsky BH. The genotype of the original Wiskott phenotype. N Engl J Med. 2006;355(17):1790–3.
Tsuboi S, Meerloo J. Wiskott-Aldrich syndrome protein is a key regulator of the phagocytic cup formation in macrophages. J Biol Chem. 2007;282(47):34194–203.
Lorenzi R, Brickell PM, Katz DR, Kinnon C, Thrasher AJ. Wiskott-Aldrich syndrome protein is necessary for efficient IgG-mediated phagocytosis. Blood. 2000;95(9):2943–6.
Greer WL, Shehabeldin A, Schulman J, Junker A, Siminovitch KA. Identification of WASP mutations, mutation hotspots and genotype-phenotype disparities in 24 patients with the Wiskott-Aldrich syndrome. Hum Genet. 1996;98(6):685–90.
Standen GR, Lillicrap DP, Matthews N, Bloom AL. Inherited thrombocytopenia, elevated serum IgA and renal disease: identification as a variant of the Wiskott-Aldrich syndrome. Q J Med. 1986;59(228):401–8.
Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–60.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164.
Yin HL, Stull JT. Proteins that regulate dynamic actin remodeling in response to membrane signaling minireview series. J Biol Chem. 1999;274(46):32529–30.
Haddad E, Zugaza JL, Louache F, Debili N, Crouin C, Schwarz K, Fischer A, Vainchenker W, Bertoglio J. The interaction between Cdc42 and WASP is required for SDF-1-induced T-lymphocyte chemotaxis. Blood. 2001;97(1):33–8.
Kolluri R, Tolias KF, Carpenter CL, Rosen FS, Kirchhausen T. Direct interaction of the Wiskott-Aldrich syndrome protein with the GTPase Cdc42. Proc Natl Acad Sci U S A. 1996;93(11):5615–8.
Snapper SB, Meelu P, Nguyen D, Stockton BM, Bozza P, Alt FW, Rosen FS, von Andrian UH, Klein C. WASP deficiency leads to global defects of directed leukocyte migration in vitro and in vivo. J Leukoc Biol. 2005;77(6):993–8.
Westerberg L, Larsson M, Hardy SJ, Fernandez C, Thrasher AJ, Severinson E. Wiskott-Aldrich syndrome protein deficiency leads to reduced B-cell adhesion, migration, and homing, and a delayed humoral immune response. Blood. 2005;105(3):1144–52.
Acknowledgements
The authors also gratefully acknowledge the patients and their family for their participation in this research study.
Funding
This work was supported by Dr. Mohammad Ali Faghihi. This work was partly supported by the US NIH NINDS R01NS081208-01A1 awarded to Mohammad Ali Faghihi. Additionally, this work was partly supported by the NIMAD research grant (940714) awarded to Mohammad Ali Faghihi. The funding agencies has no role in the design of the study and collection, analysis, and interpretation of data.
Availability of data and materials
All data from this study that do not pertain to identifiable patient information are freely available and provided as supplemental material and/or can be provided by contacting corresponding author. The identified mutation has been uploaded into HGMD database as well as free ClinVar website.
Author information
Authors and Affiliations
Contributions
MAF conceived and designed the study, collected, assembled, interpreted NGS data and wrote the manuscript. HE, MRB, AA and AK clinically evaluated the patients, wrote the case presentation and edited the manuscript. HD wrote the manuscript, designed WAS primers, interpreted Sanger sequencing results and bioinformatics analysis of WAS gene. MS performed whole exome sequencing. MAFF analyzed the NGS data, FS and ZT performed Sanger sequencing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethic committee at Shiraz University of Medical Sciences (SUMS) has approved the study and parent of affected individual has signed written consent indicating their voluntary contribution to the current study and consent to publication. A copy of the consent is available for review by the Editor of this journal.
Consent for publication
The parent of the patient consented to the publication of the case and any accompanying images with written consent.
Competing interests
The authors declare that there are no financial and non-financial competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Esmaeilzadeh, H., Bordbar, M.R., Dastsooz, H. et al. A novel splice site mutation in WAS gene in patient with Wiskott-Aldrich syndrome and chronic colitis: a case report. BMC Med Genet 19, 123 (2018). https://doi.org/10.1186/s12881-018-0647-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-018-0647-0