Abstract
Background
Many published studies have estimated the association between the +331G/A (rs10895068) polymorphism in the progesterone receptor (PgR) gene and breast cancer risk. However, the results remain inconsistent and controversial. To address this inconsistency, we systematically interrogated the aforementioned association via a meta-analysis.
Methods
Through a literature search, we identified 13 case-control studies, including 12,453 cases and 14,056 case-free controls. The strengths of reported associations were evaluated using odds ratios (ORs) with 95% confidence intervals (95%CIs).
Results
An association was found between +331G/A polymorphism and +331G/A risk in the dominant model (p = 0.027). Via subgroup analysis, we found no association between +331G/A and breast cancer risk in Caucasians, Asians or mixed racial groups.
Conclusions
Through meta-analysis, we were able to gain insight into previously reported associations between +331G/A polymorphism and breast cancer risk. However, further studies are still needed to provide more evidence.
Similar content being viewed by others
Background
The most common malignant neoplasm in women, breast cancer has a higher developed versus developing countries. It is a complex and multi-factorial disease caused by a combination of genetic and environmental factors. Although the exact mechanism of breast cancer carcinogenesis is still not completely elucidated, many factors are known to influence its development including age, nulliparity, early menarche, late menopause, and family history [1]. In addition, inherited susceptibility accounts for approximately 27% of breast cancer risk, demonstrating that genetic factors contribute to risk of developing breast cancer [2].
Progesterone (PR) is known to regulate cell proliferation and differentiation in the female reproduction system [3]. Dysregulated oestrogen and progesterone signaling results in disorders such as breast cancer, subfertility, endometriosis, and endometrial cancer that depend on steroid hormones [4]. Negative associations between PR protein levels and pathological grade, tumor size, and axillary lymph node involvement are frequently reported [5,6,7,8]. Additionally, PR positive tumors are believed to confer a more favorable prognosis. Moreover, primary breast tumors which lack PR are more prone to develop secondary sites than tumors which express PR in those postmenopausal patients [9]. This suggests that PR may also limit breast cancer progression.
The progesterone receptor (PgR) is essential for mediating the effects of progesterone, which is necessary to establish and maintain pregnancy. The PgR gene encodes two iso-forms, PR-A and PR-B. Breast cancers commonly express a predominance of one PR isoform, and the loss of coordinated expression in the ratio between PR-A/PR-B proteins within a cell is likely to result in an aberrant hormonal response [10]. The PgR gene contains eight exons and seven introns (A-G), and is located on chromosome 11q22–23 [11]. While associations between PgR gene polymorphisms and breast cancer have been well-studied, results remain inconsistent [12,13,14,15].
Among the variations of PgR gene, the +331G/A variant (rs10895068), locating in the promoter region, has been wildly studied. One case–control study including 990 cases and 1364 controls showed that the +331G > A polymorphism increases PR-B isoform expression, which is reported to increase PR-B-dependent mammary cell proliferation, thereby promoting breast cancer [16]. However, no association was found between +331G > A and breast cancer risk in a recent study of postmenopausal women [14]. Although a biological mechanism is plausible, the role of the +331G > A polymorphism in breast cancer remains ambiguous. We hypothesize that conflicting results are due to the limited sample sizes as well as differing genetic backgrounds. Meta-analysis can be used as a statistical method to reconcile studies with inconsistent results [17]. Therefore, we employed this method to investigate the relationship between the PrG +331G/A polymorphism and breast cancer risk.
Methods
Selection of eligible studies
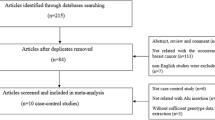
We used four online electronic databases to select studies to include in this meta-analysis (PubMed, Web of Science, and Embase in English and China National Knowledge Infrastructure Database in Chinese; most recent search update, February 2017). Search terms included “breast cancer” or “breast neoplasm” or “mammary” combined with “progesterone receptor gene” or “PgR” or “+331G/A” or “+331G > A” or “rs10895068” and with “polymorphism” or “variant” or “genotype” or “allele”, without any limitation applied. Referenced lists of all included studies were then manually searched to identify any additional eligible studies. Only the study with the most recent, complete data was included when multiple studies included the same set of subjects.
Inclusion and exclusion criteria
Included studies met the following criteria: (1) case-control design; (2) clinical trial evaluating associations between +331G/A gene polymorphisms and breast cancer susceptibility; (3) pathological confirmation of breast cancer diagnosis was reported for all patients; (4) data regarding sample size and individual genotype frequencies were available for all cases and controls; and (5) at least two comparison groups (cancer group and control group) were included. Exclusion criteria: (1) duplication of prior studies and (2) meta-analysis, letters, reviews, or editorial articles.
Data extraction
Two investigators (Xing-ling Qi and Jun Yao) independently extracted data from eligible studies. Inconsistencies were resolved via discussion between the investigators. We recorded the first author’s name, publication year, country of origin, ethnicity studied, sample size, genotypes and allele frequencies for patients with the PgR +331G/A polymorphism, and Hardy-Weinberg equilibrium (HWE) results for controls groups. We recorded studies including more than one ethnicity as mixed ethnicity.
Statistical analysis
We used STATA 12.0 software (Stata Statistical software, College Station, TX, USA) to perform all statistical analysis. PRISMA checklists and guidelines were adhered to when performing the meta-analysis [18]. For control groups, we used Chi-square tests to analyze the Hardy-Weinberg equilibrium (HWE), with p < 0.05 indicating a significant deviation. Pooled frequency analyses were performed using Thakkinstian’s method [18, 19]. The strength of associations between the +331G/A polymorphism and breast cancer risk were evaluated using odds ratios (ORs) and 95% confidence intervals (CIs). Two-tailed tests were used to generate all p values.
We used five models to evaluate associations the +331G/A and breast cancer risk: allele model, dominant model, recessive model, homozygote comparison model, and heterozygote model. A random effects model was used to pool effect sizes of all included studies for a possible effect size across populations with different genetic backgrounds after considering the heterogeneity among the included studies [20]. We also used A as the risk allele to compare OR1 (AA vs. aa), OR2 (Aa vs. aa), and OR3 (AA vs. Aa) and further determined the genetic model that was the most appropriate under the instruction, as previously described [21, 22]. Heterogeneities among studies were estimated using an I2 test, and describe I2 values as low (25%), moderate (50%), or high (75%) estimates [22]. A Z-test resulting in a p value less than 0.05 determined statistical significance. We also explored the effect of included studies on combined ORs via sensitivity analysis employing sequential omission of each study. In addition, we conducted subgroup analyses by ethnicity (i.e. Caucasian, Asian, and mixed races) as well as by source of control subjects (i.e. hospital-based vs. population-based). We generated a funnel plot to reflect any possible publication bias [23, 24], with an Egger’s test resulting in ap < 0.05 indicating significant publication bias.
Results
We performed the online search of multiple databases for available studies reporting associations between PgR +331G/A polymorphisms and breast cancer risk. We included 13 original articles in this meta-analysis after meeting inclusion criteria. As shown in Table 1, the studies eventually involved 12,453 patient and 14,056 control subjects [12, 14, 16, 25,26,27,28,29,30,31,32,33]. The frequencies of each genotype and allele along with their HWE values were described in Table 2. All studies reported control genotype distributions in accordance with HWE, save for that of Kotsopoulos, et al. (2009) (p < 0.0001) [25].
Association between PgR +331G > A and breast cancer
Table 3 shows our results generated using five genetic models to evaluate associations between the +331G > A polymorphism and breast cancer risk. Genetic model selection principles were used to determine the dominant model. Our summary results indicate that an association is indeed present between PgR +331G > A and the risk of breast cancer. Using a random effects model, we calculated a pooled OR of 1.140 (p = 0.027, 95% CI = 1.015–1.279) (Fig. 1).
Subgroup analysis
We found no association between +331G/A polymorphism and breast cancer risk in Caucasian (p = 0.102, OR = 1.116, 95% CI = 0.978–1.272,), Asian (p = 0.105, OR = 1.749, 95% CI = 0.890–3.438) and mixed race (p = 0.231, OR = 1.170, 95% CI = 0.905–1.513) populations via subgroup analysis. Furthermore, using subgroup analysis by source of controls, there was an association between +331G/A locus and breast cancer risk in hospital-based (p = 0.004, OR = 1.295, 95% CI = 1.087–1.543,), but not in population-based controls (p = 0.440, OR = 1.046, 95% CI = 0.934–1.171; Table 4).
Sensitivity analysis
We examined the influence of individual studies the pooled ORs for +331G/A via sensitivity analysis involving omitting each study in each genetic model; the results did not change. This indicates that our results are statistically robust for all five genetic models examining associations between +331G/A and breast cancer susceptibility.
Publication bias
We assessed possible publication bias using a Begg’s funnel plot and Egger’s test. As shown in Fig. 2, no obvious asymmetry was observed in the funnel plot all genotypes in the overall population, and Begg’s test results did not reveal any publication bias (p > 0.05).
Discussion
This meta-analysis included 12,453 breast cancer cases and 14,056 controls, and was used to evaluate reported associations between breast cancer risk and the +331G/A (rs10895068) functional polymorphism in the PgR gene promoter. In the dominant model, when all studies meeting eligibility criteria were pooled, we found an association between +331G/A and breast cancer risk. However, after subgroup analysis, this association disappeared in Caucasians, Asian, and mixed race. Therefore, we could cautiously assert that there is no association of the +331G/A PgR gene polymorphism and breast cancer susceptibility in Caucasian and Asian populations.
There have been several prior meta-analysis studies reporting on this particular association, with mixed results. An association between breast cancer risk and PgR +331G/A was reported by Yang, et al. [34]. However, the other two published meta-analyses, which each included more studies than that of Yang, et al., did not confirm this association [35, 36]. The present study, however, has several advantages over these prior studies. First, more recently-published studies were included in the present meta-analysis, which may underscore the reliability of our findings. Second, the present study added additional subgroup analyses by both ethnicity and sources of controls to control for heterogeneity. Third, we also included a Chinese database in our literature search to more comprehensively assess studies in Chinese populations. These advantages allowed us to more precisely assess the + 331G/A PgR gene polymorphism and breast cancer risk associations than previous meta-analyses.
There were several limitations to this study that may have affected our results. Firs, only 13 studies were included in our meta-analysis, which limited subsequent analyses because of a shortage of original studies. Second, there was moderate heterogeneity in the overall meta-analysis and in the subgroup analysis that suggested that ethnicity and source of controls, to some extent, contributed heterogeneity between studies. Third, other factors influencing breast cancer, such as genetic background, environment, and lifestyle factors, should also be considered. Finally, there was only ten studies that specified Caucasians and just one study that compared certain populations (Asian and mixed race) in the ethnicity sub-group analyses. Thus, the discrepancy of association among different ethnic sub-groups should be interpreted carefully.
Conclusion
In conclusion, our meta-analysis suggested that the +331G/A polymorphism may not be associated with susceptibility to breast cancer. However, because of the comparatively insufficient number of published studies included, our conclusions require support from additional studies. More evidence from epidemiologic studies is required to validate our results regarding the role of +331G/A (rs10895068) in the genetic susceptibility to breast cancer.
Abbreviations
- CIs:
-
confidence interval
- HWE:
-
Hardy-Weinberg equilibrium
- ORs:
-
Odds ratios
- PgR:
-
progesterone receptor
- PR:
-
Progesterone
References
Dumitrescu RG, Cotarla I. Understanding breast cancer risk -- where do we stand in 2005? J Cell Mol Med. 2005;9(1):208–21.
O'Brien JM. Environmental and heritable factors in the causation of cancer: analyses of cohorts of twins from Sweden, Denmark, and Finland, by P. Lichtenstein, N.V. Holm, P.K. Verkasalo, A. Iliadou, J. Kaprio, M. Koskenvuo, E. Pukkala, A. Skytthe, and K. Hemminki. N Engl J Med 343:78-84, 2000. Survey of ophthalmology. 2000;45(2):167–8.
Clarke CL, Sutherland RL. Progestin regulation of cellular proliferation. Endocr Rev. 1990;11(2):266–301.
Romano A, Delvoux B, Fischer DC, Groothuis P. The PROGINS polymorphism of the human progesterone receptor diminishes the response to progesterone. J Mol Endocrinol. 2007;38(1–2):331–50.
Stonelake PS, Baker PG, Gillespie WM, Dunn JA, Spooner D, Morrison JM, Bundred NJ, Oates GD, Lee MJ, Neoptolemos JP, et al. Steroid receptors, pS2 and cathepsin D in early clinically node-negative breast cancer. Eur J Cancer. 1994;30A(1):5–11.
Alghanem AA, Hussain S. The effect of tumor size and axillary lymph node metastasis on estrogen and progesterone receptors in primary breast cancer. J Surg Oncol. 1986;31(3):218–21.
Silva JS, Cox CE, Wells SA Jr, Paull D, Dilley WG, McCarty KS Sr, Fetter BF, Glaubitz LC, McCarty KS Jr. Biochemical correlates of morphologic differentiation in human breast cancer. Surgery. 1982;92(3):443–9.
Stal O, Brisfors A, Carstensen J, Ferraud L, Hatschek T, Nordenskjold B. Relationships of DNA ploidy, S-phase fraction and hormone receptor status to tumor stage in breast cancers detected by population screening. The south-East Sweden breast cancer group. Int J Cancer. 1992;51(1):28–33.
Balleine RL, Earl MJ, Greenberg ML, Clarke CL. Absence of progesterone receptor associated with secondary breast cancer in postmenopausal women. Br J Cancer. 1999;79(9–10):1564–71.
Mote PA, Bartow S, Tran N, Clarke CL. Loss of co-ordinate expression of progesterone receptors a and B is an early event in breast carcinogenesis. Breast Cancer Res Treat. 2002;72(2):163–72.
Rousseau-Merck MF, Misrahi M, Loosfelt H, Milgrom E, Berger R. Localization of the human progesterone receptor gene to chromosome 11q22-q23. Hum Genet. 1987;77(3):280–2.
Johnatty SE, Spurdle AB, Beesley J, Chen X, Hopper JL, Duffy DL, Chenevix-Trench G. Kathleen Cuningham consortium for research in familial breast C: progesterone receptor polymorphisms and risk of breast cancer: results from two Australian breast cancer studies. Breast Cancer Res Treat. 2008;109(1):91–9.
Fabjani G, Tong D, Czerwenka K, Schuster E, Speiser P, Leodolter S, Zeillinger R. Human progesterone receptor gene polymorphism PROGINS and risk for breast cancer in Austrian women. Breast Cancer Res Treat. 2002;72(2):131–7.
Feigelson HS, Rodriguez C, Jacobs EJ, Diver WR, Thun MJ, Calle EE. No association between the progesterone receptor gene +331G/a polymorphism and breast cancer. Cancer Epidemiol Biomarkers Prev. 2004;13(6):1084–5.
Gabriel CA, Mitra N, Demichele A, Rebbeck T. Association of progesterone receptor gene (PGR) variants and breast cancer risk in African American women. Breast Cancer Res Treat. 2013;139(3):833–43.
De Vivo I, Hankinson SE, Colditz GA, Hunter DJ. A functional polymorphism in the progesterone receptor gene is associated with an increase in breast cancer risk. Cancer Res. 2003;63(17):5236–8.
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–9. W264
Thakkinstian A, McEvoy M, Minelli C, Gibson P, Hancox B, Duffy D, Thompson J, Hall I, Kaufman J, Leung TF, et al. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol. 2005;162(3):201–11.
Yao J, Pan YQ, Ding M, Pang H, Wang BJ. Association between DRD2 (rs1799732 and rs1801028) and ANKK1 (rs1800497) polymorphisms and schizophrenia: a meta-analysis. Am J Med Genet Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatr Genet. 2015;168B(1):1–13.
Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends in genetics : TIG. 2004;20(9):439–44.
Thakkinstian A, McElduff P, D'Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24(9):1291–306.
Wu MS, Huang SP, Chang YT, Shun CT, Chang MC, Lin MT, Wang HP, Lin JT. Tumor necrosis factor-alpha and interleukin-10 promoter polymorphisms in Epstein-Barr virus-associated gastric carcinoma. J Infect Dis. 2002;185(1):106–9.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295(6):676–80.
Kotsopoulos J, Tworoger SS, De Vivo I, Hankinson SE, Hunter DJ, Willett WC, Chen WY. +331G/a variant in the progesterone receptor gene, postmenopausal hormone use and risk of breast cancer. Int J Cancer. 2009;125(7):1685–91.
Romano A, Lindsey PJ, Fischer DC, Delvoux B, Paulussen AD, Janssen RG, Kieback DG. Two functionally relevant polymorphisms in the human progesterone receptor gene (+331 G/a and progins) and the predisposition for breast and/or ovarian cancer. Gynecol Oncol. 2006;101(2):287–95.
Pooley KA, Healey CS, Smith PL, Pharoah PD, Thompson D, Tee L, West J, Jordan C, Easton DF, Ponder BA, et al. Association of the progesterone receptor gene with breast cancer risk: a single-nucleotide polymorphism tagging approach. Cancer Epidemiol Biomarkers Prev. 2006;15(4):675–82.
Jin YL, Shen YP, Chen ea JL. A case-control study on the associations of ER codon 325 and PR +331G/a with the risk of breast cancer. Tumor. 2008;28(10):859–63.
Fernandez LP, Milne RL, Barroso E, Cuadros M, Arias JI, Ruibal A, Benitez J, Ribas G. Estrogen and progesterone receptor gene polymorphisms and sporadic breast cancer risk: a Spanish case-control study. Int J Cancer. 2006;119(2):467–71.
al ARMBHMe. Impact of two functional progesterone receptor polymorphisms (PRP): +331G/a and PROGINS on the cancer risks in familial breast/ovarian cancer. The Open Cancer Journal. 2007;1:1–8.
Reding KW, Li CI, Weiss NS, Chen C, Carlson CS, Duggan D, Thummel KE, Daling JR, Malone KE. Genetic variation in the progesterone receptor and metabolism pathways and hormone therapy in relation to breast cancer risk. Am J Epidemiol. 2009;170(10):1241–9.
Huggins GS, Wong JY, Hankinson SE, De Vivo I. GATA5 activation of the progesterone receptor gene promoter in breast cancer cells is influenced by the +331G/a polymorphism. Cancer Res. 2006;66(3):1384–90.
Diergaarde B, Potter JD, Jupe ER, Manjeshwar S, Shimasaki CD, Pugh TW, Defreese DC, Gramling BA, Evans I, White E. Polymorphisms in genes involved in sex hormone metabolism, estrogen plus progestin hormone therapy use, and risk of postmenopausal breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(7):1751–9.
Yang DS, Sung HJ, Woo OH, Park KH, Woo SU, Kim AR, Lee ES, Lee JB, Kim YH, Kim JS, et al. Association of a progesterone receptor gene +331 G/a polymorphism with breast cancer risk: a meta-analysis. Cancer Genet Cytogenet. 2010;196(2):194–7.
KD Y, Chen AX, Shao ZM. No association between a progesterone receptor gene promoter polymorphism (+331G>A) and breast cancer risk in Caucasian women: evidence from a literature-based meta-analysis. Breast Cancer Res Treat. 2010;122(3):853–8.
Chaudhary S, Panda AK, Mishra DR, Mishra SK. Association of +331G/a PgR polymorphism with susceptibility to female reproductive cancer: evidence from a meta-analysis. PLoS One. 2013;8(1):e53308.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
YZ was in charge of conceived and designed the study. XLQ and JY were responsible for collection of data and performing the statistical analysis and manuscript preparation. YZ and XLQ were responsible for checking the data. All authors were responsible for drafting the manuscript, read and approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Qi, Xl., Yao, J. & Zhang, Y. No association between the progesterone receptor gene polymorphism (+331G/a) and the risk of breast cancer: an updated meta-analysis. BMC Med Genet 18, 123 (2017). https://doi.org/10.1186/s12881-017-0487-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-017-0487-3