Abstract
Background
Streptococcus agalactiae (Group B Streptococcus, GBS) serotypes, sequence types, and antimicrobial resistance profile vary across different geographic locations affecting disease patterns in newborns. These differences are important considerations for vaccine development efforts and data from large countries in Africa is limited. The aim of this study was to determine serotypes and genotypes of GBS isolates from pregnant women and their newborns in Ethiopia.
Methods
A hospital based cross-sectional study was conducted at three hospitals in Ethiopia from June 2014 to September 2015. Out of 225 GBS isolates, 121 GBS were recovered, confirmed and characterized at CDC’s Streptococcus Laboratory using conventional microbiology methods and whole genome sequencing.
Results
Of the 121 isolates, 87 were from rectovaginal samples of pregnant women, 32 from different body parts of their newborns and 2 from blood of newborns with suspected sepsis. There were 25 mother-infant pairs and 24 pairs had concordant strains. The most prevalent serotypes among mothers and/or their babies were II, Ia and V (41.5, 20.6, 19.5 and 40.6%, 25 and 15.6%, respectively). Multilocus sequence typing (MLST) on 83 isolates showed ST10 (24; 28.9%) and ST2 (12; 14.5%) as most predominant sequence types. All GBS strains were susceptible to penicillin, cefotaxime and vancomycin, which correlated to the presence of wildtype PBP2x types and the lack of known vancomycin-resistance genes. Tetracycline resistance was high (73; 88%, associated primarily with tetM, but also tetO and tetL). Five isolates (6%) were resistant to erythromycin and clindamycin and 3 isolates were fluoroquinolone-resistant, containing associated mutations in gyrA and parC genes. All isolates were positive for one of four homologous Alpha/Rib family determinants and 1–2 of the three main pilus types.
Conclusions
Predominant serotypes were II, Ia, and V. A limited number of clonal types were identified with two STs accounting for about half of the isolates. All strains collected in this study were susceptible to beta-lactam antibiotics and vancomycin. Typical of most GBS, these isolates were positive for single alpha-like family protein, serine-rich repeat gene, as well as 1–2 pilus determinants.
Similar content being viewed by others
Background
Group B Streptococcus (GBS) is a recognized cause of infant sepsis and meningitis globally and is a leading cause of morbidity and mortality in Africa [1]. GBS is also a common commensal colonizer of the gastrointestinal and urogenital tracts of women and maternal colonization is a major factor in mother-to-child GBS transmission. Early onset (< 7 days) GBS disease has been well characterized, whereas the epidemiology of late onset disease (LOD, 7–89 days) is less well understood [2].
Penicillin G is the drug of choice for intrapartum prophylaxis [3] and GBS isolates remain mostly susceptible to penicillin. Its prophylactic use has been instrumental in significantly reducing the incidence of early-onset diseases in neonates [4]. However, GBS isolates with reduced susceptibility to penicillin have been reported for more than a decade [5]. Erythromycin and clindamycin have been used as alternatives to prevent vertical transmission of GBS among pregnant women who are allergic to penicillin, but resistance to these antibiotics has emerged in several countries, including reports from Africa [6, 7].
GBS strains are subdivided according to type-specific capsular polysaccharides into 10 unique serotypes which are also a major focus of vaccine development [8]. Serotypes I to V account for about 98% of colonizing GBS isolates worldwide with serotype III usually associated with invasive disease and less common among colonization isolates. GBS serotype distribution is not uniform across different geographic regions and temporal variations have also been described [9]. As information on GBS from their genomes has accumulated, molecular methods have also proved very useful for investigating the population structure of GBS and discriminating differences between strains isolated from different sources [5, 10,11,12]. For example, the ST17 lineage has been associated with neonatal infections, particularly with late-onset disease [11]. There are previous reports showing diversification and shifts in serotypes [5, 12, 13] and abundant evidence of past capsular switching within several MLST-based lineages [12, 14] which also pose potential challenges for the development of a vaccine. Global data on circulating GBS strains is important for disease control and for informing the development of effective vaccines. However relatively few studies are available from Sub-Saharan Africa [1] and there are limited data on serotypes and strain characterization [7].
Ethiopia is an important country with a substantial birth cohort in Africa, and while there are previous data published from Ethiopia on antibiotic susceptibility patterns for GBS, there are limited available detailed descriptive data on circulating strains [7, 15]. Here we used whole genome sequencing to investigate serotype distribution, clonal relationships, lineage distributions, virulence factor determinants, and antimicrobial susceptibility patterns of S. agalactiae strains recovered from pregnant mothers and their newborns attending three hospitals in Ethiopia.
Methods
Study population
This prospective, cross-sectional study was conducted at 3 hospitals in Ethiopia: Adama Hospital Medical College (AHMC), Hawassa University Comprehensive Specialized Hospital (HUCSH) and Tikur Anbessa Specialized Hospital (TASH) between June 2014 and September 2015. The three hospitals were selected based on their convenience. AHMC is a rural hospital located in Adama City in the Oromia regional state. It is located 100 km due east of Addis Ababa and has a total population of 220,212. HUCSH, a rural hospital, is located in Hawassa which is the capital city of Southern Nations Nationalities and Peoples Region (SNNRP) and is located 275 km south of Addis Ababa. The total population of Hawassa town is 235,000. TASH is an urban hospital located in Addis Ababa, the capital city of Ethiopia, with a large population size of 3,384,569.
Eligibility criteria
Pregnant women who were admitted for delivery along with their newborn were included. Pregnant women with cesarean section delivery and those who were on antibiotic treatment for the last 2 weeks prior to data collection were excluded.
Newborns who were suspected of neonatal disease, those with signs and symptoms of neonatal disease (breathing problem, reduced movement, reduced suckling, seizure, slow or increased heart rate, vomiting, increased or reduced body temperature).
Isolation of Bacteria
Recto-vaginal swabs from 840 pregnant women, samples from the nasal area, external ear, umbilical cord or throat area of 857 newborns and blood samples from newborns suspected of neonatal disease were collected. The detail are as follows: Number of pregnant women and their newborn at AHMC, Adama were 280 and 282 respectively; data collection period was from June 2014 to October 2014. Number of pregnant women and their newborn at HUCSH, Hawassa were 280 and 292 respectively; data collection period was November 2014 to March 2014. Number of pregnant women and their newborn at TASH, Addis Ababa were 280 and 283 respectively; data collection period was March 2015 to August 2015. One hundred seventy-six newborns suspected of early onset disease (defined as occurring during the first week of life) were included from TASH, Addis Ababa; data collection period was from March 2015 to August 2015.
At each study site, to isolate GBS from pregnant women and newborns, recto-vaginal swabs from mothers and samples from the nasal area, external ear, umbilical cord or throat area of newborns were placed into Lim broth (BD Diagnostics, USA) and incubated for 18–24 h at 37 °C in CO2 enriched atmosphere. Then sub-cultured onto sheep blood agar (BD Diagnostics, USA) and incubated in CO2 enriched atmosphere at 37 °C for 18–24 h. If there was no growth, blood agar plate was re-incubated and examined after 48 h.
To isolate GBS from newborns suspected of early onset disease, about 1 ml blood was inoculated into Tryptone Soy Broth (BD Diagnostics, USA). All blood cultures were incubated aerobically at 37 °C and inspected daily for 7 days for the presence of visible microbial growth by observing any of one of the following changes: turbidity, hemolysis, gas production and coagulation of broth. Blood cultures with sign of microbial growth were sub-cultured onto blood agar (BD Diagnostics, USA). The blood agar plate was incubated aerobically in CO2 enriched atmosphere at 37 °C for 24–48 h.
To identify GBS, hemolytic reaction on BAP (beta-hemolytic or non-hemolytic), Gram reaction, catalase test, CAMP (Christie, Atkins, and Munch-Petersen) test and Strep B Grouping Latex (Remel, USA) were used. Isolates were stored at − 70 °C in medium containing skim milk, tryptone, glucose, and glycerol [16] and transported to the Streptococcus Laboratory at the Centers for Disease Control and Prevention for confirmation and characterization. For several analyses, where more than one isolate was available for mothers and/or babies and they had the same serotype and MLST type, only a single isolate was selected so as not to duplicate results.
DNA extraction and whole genome sequencing
At CDC, isolates were cultured on Trypticase soy agar supplemented with 5% sheep blood (BAP). A positive CAMP test and Strep. B Grouping Latex (Remel, USA) were used to confirm isolates as S. agalactiae.
For whole genome sequencing (WGS), GBS isolates were cultured on BAPs and incubated overnight at 37 °C. Genomic DNA was extracted manually using a modified QIAamp DNA mini kit protocol (Qiagen, Inc., Valenica, CA, USA) (https://www.cdc.gov/streplab/downloads/pcr-body-fluid-dna-extract-strep.pdf). Nucleic acid concentration was quantified by Qubit assay (Thermo Fisher Scientific Inc., Waltham, MA, USA) and samples were sheared using a Covaris M220 ultrasonicator (Covaris, Inc., Woburn, MA, USA) programmed to generate 500-bp fragments. Libraries were constructed on the SciCloneG3 (PerkinElmer Inc., Waltham, MA, USA) using an Ovation Rapid multiplex library preparation kit with 96 dual indices (NuGEN, San Carlos, CA, USA) and quantified by KAPA qPCR library quantification method (Kapa Biosystem Inc., Wilmington, MA, USA). Short read sequences were generated with MiSeq v2 500 cycle kit (Illumina Inc). Isolate identifiers, pipeline features, and assembly metrics are listed in Supplementary Table 1. For the 119 isolates that yielded high quality sequencing metrics with contig size below 500, sequences are available in the NCBI repository and accession number provided in Supplemental Table 1.
Serotyping and surface protein detection
A multiplex PCR assay was initially used for the direct identification of the capsular serotypes (Ia to IX) of GBS [17]. In addition, serotype was confirmed using the CDC short-read WGS bioinformatics pipeline [5, 12]. The presence of hypervirulent GBS adhesion determinant (hvgA), serine-rich repeat gene (srr), one of four alpha-family surface protein genes (rib, alpha, alp1, alp2/3) and the pilus islands (PI-1, PI-2a, and PI-2b) were also queried through the CDC pipeline (https://github.com/BenJamesMetcalf) [12].
Multilocus sequence typing
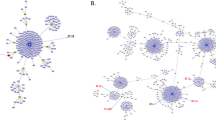
Seven locus MLST to assign sequence type (ST) was facilitated from whole genome sequence data with the CDC pipeline using SRST2 and database at http://pubmlst.org/sagalactiae/ [5]. eBURST was used to group isolates into lineages or clonal complexes (CCs), based upon sharing at least six of 7 MLST alleles with one or more other members [18]. The relations between STs and serotypes of GBS isolates were illustrated by the minimum spanning tree (PHYLOVIZ software version 2.0; PHYLOViZ team, Lisbon, Portugal).
Antimicrobial susceptibility testing
Antimicrobial susceptibility patterns of GBS were tested by broth microdilution. The antimicrobials tested included penicillin, cefotaxime, erythromycin, clindamycin, levofloxacin, vancomycin, daptomycin, tetracycline, and linezolid. Isolates were classified as sensitive, intermediate, or resistant according to Clinical Laboratory Standards Institute (CLSI) guidelines [19]. Strains were determined to be multidrug resistant if resistant to ≥3 different antibiotic classes. Phenotypic MICs were compared with WGS-predictions for non-β-lactam antibiotics (except daptomycin) using sequence queries and a bioinformatics pipeline (https://github.com/BenJamesMetcalf/Spn_Scripts_Reference) for detection of resistance determinants provided in a previous study [5]. The PBP2x typing scheme used serves to flag missense mutations within the pbp2x gene for subsequent isolate MIC testing for beta-lactam antibiotics.
Results
Source, serotype and MLST profiles of S. agalactaie
Among 225 S. agalactiae isolates collected at the 3 study sites, only 121 isolates were recovered and confirmed as GBS at CDC’s Streptococcus Laboratory for further characterization. Of these 121 GBS isolates, 87 were from rectovaginal samples from healthy pregnant women, 32 from different body parts of their healthy newborns (ear, throat, and nasal), and 2 from newborns with suspected early-onset disease. Twenty-eight isolates (22.8%) were collected at AHMC, Oromia Regional state (June 2014 to October 2014); 60 (48.8%) at HUCSH, Sidama Regional State (November 2014 to March 2014) and 33 (27.3%) were from TASH, Addis Ababa, the capital city of Ethiopia (March 2015 to August 2015). Of the 121 isolates, there were 25 mother-infant pairs and nine cases with > 1 isolate for mother or baby. A total of 83 non-duplicated isolates were identified for analysis of antimicrobial susceptibility and genotypes.
Six of the ten known serotypes occurred in this study population: For mother isolates: II (n = 36; 41.5%), Ia (n = 18; 20.6%), V (n = 17; 19.5%), Ib (n = 11; 12.6%), and III (n = 5; 5.7%) and for baby colonization isolates: II (n = 13; 40.6%), Ia (n = 8; 25%), V (n = 5; 15.6%), III (n = 4; 12.5%), Ib (n = 3; 9.3% and IV (n = 1: 3.1%) (Fig. 1). The EOD isolates from 2 newborns were Ia (ST5) and III (ST19). There was 100% concordance with serotype prediction from PCR and the genomic prediction approach.
MLST analysis identified 17 different STs among 83 GBS strains. The predominant STs were ST10 (n = 24; 28.9%), ST2 (n = 12; 14.5%) and ST19 (n = 8; 9.6%) (Table 1). Using eBURST, the STs were grouped into 5 CCs (Fig. 2), with CC8 accounting for the majority of isolates (n = 36; 43.4%). One singleton (ST934) was identified and not part of a cluster. Most STs were associated with a single serotype except ST196 (represented by serotypes IV and V) and ST19 (represented by serotypes III and V). Serotype Ia was mostly associated with CC249 (n = 14;77.8%) and serotypes V and II with CC2 (n = 18; 100%) and CC10 (n = 24; 80%), respectively.
Surface proteins
One of the four homologous alpha protein gene family queries was detected in each isolate: alpha (37: 44.6%), alp1 (24: 28.9%), alp2/3 (7: 8.4%) or rib (15; 18.1%). The serine-rich repeat glycoprotein determinant srr1 was identified in all GBS isolates and 1–2 of the three pilus backbone determinants, each of which corresponds to a distinct pilus and pathogenicity island, were also detected in all isolates: PI-1 + PI-2a (57; 68.7%), PI-2a (11; 13.3%), PI-2b (8; 9.6%), PI-1 (7: 8.4%). Two serotype Ia/ST934 isolates (2.4%) were hvgA-positive. (Table 1).
Serotype Ia isolates were alp1-positive (61.1%) or alp2/3-positive (38.9%). All serotype Ib and III isolates were alpha-positive and rib-positive, respectively. The majority of serotype II were alpha-positive (86.7%) with 13.3% rib-positive. Similarly, most of serotype V were alp1-positive (66.7%) with a smaller number having rib determinant (33.3%) (Table 1).
Most isolates were positive for pilus subunit queries PI-1 and PI-2a (24 serotype II, 17 serotype V, 11 serotype Ib, and 5 serotype III), while seven isolates (all serotype Ia) were positive for PI-1 alone. Isolates with serotypes Ia (n = 7), II (n = 3) and V (n = 1) were PI-2a positive. The pilus subunit query for PI-2b was identified in serotype Ia (4 isolates), II (3 isolates) and in a single serotype IV isolate in the study (Table 1).
Antimicrobial susceptibility profile, resistance determinants, virulence factor determinants
All 83 non-duplicated GBS strains collected in the study were susceptible to penicillin, linezolid, cefotaxime, and vancomycin (Table 2). PBP2X types were restricted to types, 1, 4 and 5. The majority of isolates were resistant to tetracycline (n = 73; 88%) conferred by tetM (48; 57.8%), tetL + M (24; 28.9%) or tetO (1; 1.2%) (Table 1). All GBS strains of serotypes II, III, IV and V were resistant to tetracycline (Table 2). Five isolates within serotypes III and V were non-susceptible to erythromycin. These included 4 inducibly clindamycin-resistant isolates carrying ermTR and one constitutively clindamycin-resistant ermB-positive strain. Two isolates with ermTR were also positive for lnu genes. Three isolates of serotype V/ST19 had levofloxacin MICs > 4 μg/ml and had mutations within both gyrA and parC (Table 1) and these same isolates also had the aac6 (aph2) determinant that confers gentamicin resistance. Five isolates were considered MDR and this included the 3 resistant to levofloxacin.
Discussion
Vaccination of pregnant women against GBS is a promising strategy to prevent invasive GBS disease in their infants [8]. Vaccine candidates include protein-based formulations and serotype-specific polysaccharide-protein conjugates [20] and thus an understanding of serotype and surface-protein antigen distribution in maternal colonization and infant disease worldwide is important. While there have been several reports published from Ethiopia on maternal and infant GBS colonization and disease over the past decade, little information on GBS strain characteristics has been described. In this study, serotype II was the most common predicted serotype, with five types (Ia, Ib, II, III and V) accounting for the vast majority of isolates. This is consistent with maternal colonization serotypes described from other studies in Africa and globally [7, 9], however, serotype II is not usually the predominant serotype but rather serotypes III and V [9]. Globally, the vast majority of invasive and colonizing GBS isolates are grouped into five CCs (CC1, CC10, CC17, CC19, and CC23) [21, 22]. In this study, 88% of isolates were grouped into one of these CCs, although no isolates belonging to CC1 and CC17 were identified, emphasizing the diversity of S. agalactiae in human isolates and highlighting the potential for local geographic differences. A recent study from Northwest Ethiopia grouped GBS in four CCs (CC1, CC10, CC19 and CC23) [23]. Similar to findings by others [21, 22] we also saw evidence of past capsular locus replacement events within 2 genetic lineages (ST19 and ST196) which may have implications for vaccine development strategies.
Surface protein antigens play an important role in the pathogenesis of GBS infection, and several of these antigens have been documented as promising vaccine targets [24]. Our data are consistent with past work indicating that a vaccine containing the 3 pilus protein components could be effective in preventing disease caused by GBS as all strains carried at least one of the pilus proteins [25]. Similarly, in all of the isolates one of four highly related Alpha family proteins (Alpha, Rib, Alp2/3, Alp1) were detected suggesting possible broad coverage of the fused N terminal domain Rib-Alpha (GBS-NN) vaccine tested in phase I clinical trials in pregnant women [26]. The HvgA surface-anchored protein has been found to be critical for GBS intestinal colonization and translocation across the blood brain barrier during the onset of meningitis [27]. There were no isolates from meningitis cases in this study but a small proportion of GBS colonization isolates (2.4%) contained the hvgA gene. The hvgA gene has been detected in previous studies, primarily among highly virulent S. agalactiae belonging to serotype III/ST17 [27] and rarely found among non-serotype III isolates [14, 28]. Two hvgA positive isolates in our sampling were of serotype Ia/ST934, which is genetically divergent from other characterized GBS. The closest MLST match within the global database was the triple locus variant ST1232 from an invasive, hvgA-negative serotype II strain recovered during 2017 [12http://pubmlst.net].
S. agalactiae have generally been considered universally susceptible to penicillin although there have been several reports of isolates with mutations associated with decreased susceptibility over the last decade [12, 29, 30]. Here, PBP 2X types were restricted to types, 1, 4 and 5, commonly seen in penicillin-susceptible US isolates [5] and all isolates were sensitive to penicillin and cefotaxime. This supports data published from Jimma, Ethiopia [31], however, several previous colonization studies in Ethiopia have documented large numbers of GBS strains resistant to penicillin [15, 32]. Differences in these reported rates warrants further investigation to determine if these rates are indeed real or due to challenges with appropriate and accurate laboratory testing for GBS species identification and antibiotic resistance. The proportion of isolates with in vitro resistance to both erythromycin and clindamycin was similar to rates described by Mengist et al. [31] but lower than that reported from other regions of Ethiopia [7, 32] and other countries [7, 12, 33]. Combined resistance to erythromycin and clindamycin in GBS is most commonly due to 23S rRNA methylases encoded by different erm genes which supports our findings with ermTR and ermB determinants predominantly associated with macrolide and lincosamide resistance [5]. The high level of tetracycline resistance, associated with tetM resistance determinant, may strengthen the hypothesis that current globally circulating S. agalactiae strains in humans were selected by tetracycline usage in 1940s [34]. The proportion of isolates resistant to levofloxacin were similar to that from United States [5, 12], Taiwan [35], Italy [36] and Brazil [37] and lower compared to data reported from China [38] and Canada [39].
A key strength of our study was the molecular characterization of GBS isolates from a region of the world with limited data on this subject. A major challenge and limitation were the reduced recovery rate of GBS strains at CDC allowing only half of the isolates available for additional testing. The inability to recover GBS from the initial frozen stocks could partly be due to loss of viability during storage and transport, and a number of samples were also heavily contaminated which may have contributed also to reduced recovery.
Conclusions
In summary, the most prevalent GBS serotype was serotype II followed Ia, V, Ib, III and IV. This study suggests that circulating GBS in mothers and infants is primarily restricted to five major genetic lineages. All isolates were susceptible to penicillin and resistance to macrolides was relatively low. This study highlights the importance of additional studies to assess GBS epidemiology and develop accurate GBS prevention strategies in Ethiopia. Further epidemiological studies are needed for a more detailed understanding of GBS strain distributions and for subsequent development of vaccination strategies.
Availability of data and materials
WGS accessions numbers for 83 S. agalactiae isolates are as follows: SAMN14907220, SAMN14907221, SAMN14907223, SAMN14907224, SAMN14907225, SAMN14907226, SAMN14907227, SAMN14907228, SAMN14907229, SAMN14907230, SAMN14907231, SAMN14907232, SAMN14907234, SAMN14907235, SAMN14907236, SAMN14907237, SAMN14907238, SAMN14907239, SAMN14907240, SAMN14907242, SAMN14907244, SAMN14907245, SAMN14907246, SAMN14907247, SAMN14907250, SAMN14907251, SAMN14907252, SAMN14907253, SAMN14907255, SAMN14907258, SAMN14907259, SAMN14907260, SAMN14907261, SAMN14907264, SAMN14907266, SAMN14907267, SAMN14907269, SAMN14907270, SAMN14907271, SAMN14907272, SAMN14907273, SAMN14907276, SAMN14907278, SAMN14907279, SAMN14907280, SAMN14907282, SAMN14907283, SAMN14907284, SAMN14907288, SAMN14907289, SAMN14907290, SAMN14907291, SAMN14907292, SAMN14907293, SAMN14907294, SAMN14907295, SAMN14907296, SAMN14907297, SAMN14907299, SAMN14907301, SAMN14907302, SAMN14907303, SAMN14907307, SAMN14907308, SAMN14907310, SAMN14907312, SAMN14907315, SAMN14907316, SAMN14907317, SAMN14907318, SAMN14907320, SAMN14907324, SAMN14907325, SAMN14907326, SAMN14907327, SAMN14907328, SAMN14907329, SAMN14907330, SAMN14907331, SAMN14907332, SAMN14907333, SAMN14907335, SAMN14907336 (Supplemental File 1). Sequences are deposited at NCBI. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- GBS:
-
Group B Streptococcus
- CDC:
-
Centers for Disease Control and Prevention
- MLST:
-
Multilocus sequence typing
- ST:
-
Sequence type
- AHMC:
-
Adama Hospital Medical College
- HUCSH:
-
Hawassa University Comprehensive Specialized Hospital
- TASH:
-
Tikur Anbessa Specialized Hospital
- CAMP:
-
Christie, Atkins, and Munch-Petersen
- WGS:
-
Whole genome sequencing
- CC:
-
Clonal complex
- PBP:
-
Penicillin binding protein
- PCR:
-
Polymerase Chain Reaction
- HVGA:
-
Hyper Virulent GBS Adhesin
References
Sinha A, Russell LB, Tomczyk S, Verani JR, Schrag SJ, Berkley JA, et al. Disease burden of group B Streptococcus among infants in sub-Saharan Africa: a systematic literature review and meta-analysis. Pediatr Infect Dis J. 2016;35:933–42.
Seale A, Blencowe H, Bianchi-Jassir F, Embleton N, Bassat Q, Ordi J, et al. Stillbirth with group B Streptococcus disease worldwide: systematic review and meta-analyses. Clin Infect Dis. 2017;65:S125–32.
American College of Obstetricians and Gynecologists. Prevention of early-onset Group B Streptococcal disease in newborns. 2019. No. 782. https://www.acog.org/GBS.
Verani JR, Spina NL, Lynfield R, Schaffner W, Harrison LH, Holst A, et al. Early-onset group B streptococcal disease in the United States: potential for further reduction. Obstet Gynecol. 2014;123:828–37.
Metcalf BJ, Chochua S, Gertz RE Jr, Hawkins PA, Ricaldi J, Li Z, et al. Short-read whole genome sequencing for determination of antimicrobial resistance mechanisms and capsular serotypes of current invasive Streptococcus agalactiae recovered in the USA. Clin Microbiol Infect. 2017;23:574.e7–574.
Bolukaoto JY, Monyama CM, Chukwu MO, Lekala SM, Nchabeleng M, Maloba MR, et al. Antibiotic resistance of Streptococcus agalactiae isolated from pregnant women in Garankuwa, South Africa. BMC Res Notes. 2015;8:364.
Gizachew M, Tiruneh M, Moges F, Tessema B. Streptococcus agalactiae maternal colonization, antibiotic resistance and serotype profiles in Africa: a meta-analysis. Ann Clin Microbiol Antimicrob. 2019;18:14.
Heath PT. Status of vaccine research and development of vaccines for GBS. Vaccine. 2016;34:2876–9.
Russell NJ, Seale AC, O’Driscoll M, O’Sullivan C, Bianchi-Jassir F, Gonzalez-Guarin J, et al. Maternal colonization with group B Streptococcus and serotype distribution worldwide: systematic review and meta-analyses. Clin Infect Dis. 2017;65(S2):S100–11.
Jones N, Bohnsack JF, Takahashi S, Oliver KA, Chan MS. Kunst. Multilocus sequence typing system for group B Streptococcus. J Clin Microbiol. 2003;41:2530–6.
Lamy MC, Dramsi S, Billoet A, Reglier-Poupet H, Tazi A, Raymond J, et al. Rapid detection of the ‘highly virulent’ group B Streptococcus ST-17 clone. Microbes Infect. 2006;8:1714–22.
McGee L, Chochua S, Li Z, Mathis S, Rivers J, Metcalf B, Ryan A, et al. Multistate, population-based distributions of candidate vaccine targets, clonal complexes, and resistance features of invasive Group B Streptococci within the US: 2015–2017. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa151 [Epub ahead of print].
Slotved HC, Kong F, Lambertsen L, Sauer S, Gilbert GL. Serotype IX, a proposed new Streptococcus agalactiaeserotype. J Clin Microbiol. 2007;45:2929–36.
Bellais S, Six A, Fouet A, Longo M, Dmytruk N, Glaser P, et al. Capsular switching in group B streptococcus CC17 hypervirulent clone: a future challenge for polysaccharide vaccine development. J Infect Dis. 2012;206:1745–52.
Gizachew M, Tiruneh M, Moges F, Adefris M, Tigabu Z, Tessema B. Streptococcus agalactiae from Ethiopian pregnant women; prevalence, associated factors and antimicrobial resistance: alarming for prophylaxis. Ann Clin Microbiol Antimicrob. 2019;18:3.
O’Brien KL, Nohynek H. World Health Organization Pneumococcal Vaccine Trials Carriage Working Group Report from a WHO Working Group: standard method for detecting upper respiratory carriage of Streptococcus pneumoniae. Pediatr Infect Dis J. 2003;22:e1–11 Erratumin:PediatrInfectDisJ. 2011;30(2):185.
Imperi M, Pataracchia M, Alfarone G, Baldassarri L, Orefici G, Creti R. A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J Microbiol Methods. 2010;80:212–4.
Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2014;186:1518–30.
Clinical Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 24th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2014.
Heyderman RS, Madhi SA, French N, et al. Group B Streptococcus vaccination in pregnant women with or without HIV in Africa: a non-randomised phase 2, open-label, multicentre trial. Lancet Infect Dis. 2016;16:546–55.
Brzychczy-Wloch M, Gosiewski T, Bulanda M. Multilocus sequence types of invasive and colonizing neonatal group B streptococci in Poland. Med Princ Pract. 2014;23:323–30.
Strakova L, Motlova J. Active surveillance of early onset disease due to group B streptococci in new born. J Indian J med Res. 2004;119:205–7.
Gizachew M, Tiruneh M, Moges F, Adefris M, Tigabu Z, Tessema B. Molecular characterization of Streptococcus agalactiae isolated from pregnant women and newborns at the University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia. BMC Infect Dis. 2020;20:35.
Paoletti LC, Kasper DL. Surface structures of Group B Streptococcus important in human immunity. Microbiol Spectr. 2019;7(2). https://doi.org/10.1128/microbiolspec GPP3-0001-2017.
Margarit I, Rinaudo C, Galeotti C, Maione D, Ghezzo C, Buttazzoni E, et al. Preventing bacterial infections with pilus-based vaccines: the group B streptococcus paradigm. J Infect Dis. 2009;199:108–15.
Stålhammar-Carlemalm M, Waldemarsson J, Johnsson E, Areschoug T, Lindahl G. Nonimmuno dominant regions are effective as building blocks in a streptococcal fusion protein vaccine. Cell Host Microbe. 2007;2:427–34.
Tazi A, Disson O, Bellais S, Bouaboud A, Dmytruk N, Dramsi S, et al. The surface protein HvgA mediates group B streptococcus hypervirulence and meningeal tropism in neonates. J Exp Med. 2010;207:2313–22.
Teatero S, McGee A, Li A, Gomes J, Seah C, Demczuk W, et al. Population structure and antimicrobial resistance of invasive serotype IV group B Streptococcus, Toronto, Ontario, Canada. Emerg Infect Dis. 2015;21:585–91.
Kimura K, Nagano N, Arakawa Y. Classification of group B streptococci with reduced β-lactam susceptibility (GBS-RBS) based on the amino acid substitutions in PBPs. Antimicrob Agents Chemother. 2015;70:1601–3.
Yi A, Kim CK, Kimura K, Arakawa Y, Hur M, Yun YM, et al. First case in Korea of Group B Streptococcus with reduced penicillin susceptibility harboring amino acid substitutions in penicillin-binding protein 2X. Ann Lab Med. 2019;39:414–6.
Mengist A, Kannan H, Abdissa A. Prevalence and antimicrobial susceptibility pattern of anorectal and vaginal group B streptococci isolates among pregnant women in Jimma, Ethiopia. BMC Res Notes. 2016;9:351.
Assefa S, Desta K, Lema T. Group B streptococci vaginal colonization and drug susceptibility pattern among pregnant women attending in selected public antenatal care centers in Addis Ababa, Ethiopia. BMC Pregnancy and Childbirth. 2018;18:135.
Hawkins PA, Law CS, Metcalf BJ, Chochua S, Jackson DM, Westblade LF, et al. Cross-resistance to lincosamides, streptogramins a and pleuromutilins in Streptococcus agalactiae isolates from the USA. J Antimicrob Chemother. 2017;72:1886–92.
Da Cunha V, Davies MR, Douarre PE, Rosinski-Chupin I, Margarit I, Spinali S, et al. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun. 2014;5:4544.10.
Wu H-M, Janapatla R, Wu C-W, Yan J-J, Ho Y-R, Wu J-J, Hung K-H. Emergence of fluoroquinolone resistance in Group B Streptococcal isolates in Taiwan. Antimicrob Agents Chemother. 2008;52:1888–90.
Piccinelli G, Gargiulo F, Corbellini S, Ravizzola G, Bonfanti C, Caruso A, et al. Emergence of the first levofloxacin-resistant strains of Streptococcus agalactiae isolated in Italy. Antimicrob Agents Chemother. 2015;59:2466–9.
Nakamura P, Schuab R, Neves F, Pereira C, Paula C, Barros R. Antimicrobial resistance profiles and genetic characterisation of macrolide resistant isolates of Streptococcus agalactiae. Mem Inst Oswaldo Cruz. 2011;106:119–22.
Ji W, Zhang L, Guo Z, Xie S, Yang W, Chen J, et al. Colonization prevalence and antibiotic susceptibility of Group B Streptococcus in pregnant women over a 6-year period in Dongguan, China. PLoS ONE. 2017;12:e0183083.
Neemuchwala A, Teatero S, Patel S, Fittipaldi N. Fluoroquinolone resistance among clonal complex 1 group B Streptococcus strains. Canadian J Infect Dis Med Microbiol. 2016;2016:6403928.
Acknowledgments
We would like to acknowledge staff of Addis Ababa University, College of Health Science, Hawassa University, College of Medicine and Health Sciences for their valuable support during the study. We thank Benjamin Metcalf for GBS bioinformatics pipeline development and Sopio Chochua and the CDC Streptococcus Lab for genomic sequencing and assistance with bacterial species confirmation.
Funding
This study was supported by Addis Ababa University College of Health Science and Hawassa University, College of Medicine and Health Sciences. Confirmation of GBS isolates and whole genome sequencing was supported by Centers for Disease Control and Prevention Advanced Microbial Detection (AMD) project funds.
Author information
Authors and Affiliations
Contributions
MMA conceived and designed the study, performed laboratory work, data analysis and write-up. YW reviewed, supervised, analyzed results and prepared manuscript. DA reviewed, supervised and contributed to writing of manuscript. DAF performed laboratory work. LM supervised lab work at CDC, performed data analysis and was a major contributor in writing the manuscript. SS and BB provided critical review to manuscript preparation. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Institutional Review Board of the College of Health Sciences, Addis Ababa University (Ref No:069/13/DMIP) and the National Ethics and Research Committee (Ref No: 3.10/795/06). Written informed consent was obtained from all study participants to further characterize the S. agalactiae obtained from them and their newborn. CDC provided support only for characterization of blinded GBS isolates. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
Consent for publication
Not applicable.
Competing interests
The Authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Supplemental Table S1. WGS NCBI accession numbers and bioinformatics pipeline details
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ali, M.M., Woldeamanuel, Y., Asrat, D. et al. Features of Streptococcus agalactiae strains recovered from pregnant women and newborns attending different hospitals in Ethiopia. BMC Infect Dis 20, 848 (2020). https://doi.org/10.1186/s12879-020-05581-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-020-05581-8