Abstract
Background
Pandoraea species is a newly described genus, which is multidrug resistant and difficult to identify. Clinical isolates are mostly cultured from cystic fibrosis (CF) patients. CF is a rare disease in China, which makes Pandoraea a total stranger to Chinese physicians. Pandoraea genus is reported as an emerging pathogen in CF patients in most cases. However, there are few pieces of evidence that confirm Pandoraea can be more virulent in non-CF patients. The pathogenicity of Pandoraea genus is poorly understood, as well as its treatment. The incidence of Pandoraea induced infection in non-CF patients may be underestimated and it’s important to identify and understand these organisms.
Case presentation
We report a 44-years-old man who suffered from pneumonia and died eventually. Before his condition deteriorated, a Gram-negative bacilli was cultured from his sputum and identified as Pandoraea Apista by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS).
Conclusion
Pandoraea spp. is an emerging opportunistic pathogen. The incidences of Pandoraea related infection in non-CF patients may be underestimated due to the difficulty of identification. All strains of Pandoraea show multi-drug resistance and highly variable susceptibility. To better treatment, species-level identification and antibiotic susceptibility test are necessary.
Similar content being viewed by others
Background
Pandoraea species were first described by Coenye et al. in 2000 [1], which had been isolated from both environmental and human clinical samples, mostly from cystic fibrosis (CF) patients. They may contribute to the lung function decline in CF patients. Some of these organisms are capable of causing bacteremia in both CF and non-CF patients. And yet, with limited evidence presented, the pathogenicity of Pandoraea remains poorly understood. Besides, it is difficult to differentiate Pandoraea species from some other species, such as Burkholderia or Ralstonia [2]. Furthermore, these bacteria are resistant to a lot of antibiotics, which makes the treatment of Pandoraea related infections more complicated. Pandoraea is rarely found in non-CF patients, due to different lung environments and misidentification. We report a case of pneumonia caused by Pandoraea Apista after the evacuation of traumatic intracranial hematomas and review the available literature concerning Pandoraea species to a better understanding.
Case presentation
A 44-year-old man was transferred to the emergency intensive care unit (EICU) of The First Affiliated Hospital of Zhejiang University, Hangzhou, China, on November 25, 2018, due to multiple injuries and coma after a brain injury. Five days earlier, he accidentally fell from a height of about 7 m and immediately fell into a coma. Removal of traumatic intracranial hematoma and decompressive craniectomy were performed on November 20, 2018, and November 24, 2018. And antimicrobial treatment had been given before he was admitted to our hospital.
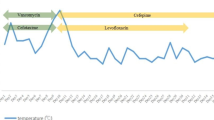
On the day of admission, physical examination showed a low-grade fever of 37.6 °C, a blood pressure of 165/79 mmHg and a Glasgow Coma Score of 1 + T + 1. Laboratory examination detected an elevated white blood cell count (17.1 × 10E9/L with 90.6% neutrophils) and hypersensitive C-reactive protein (hsCRP) of 209.70 mg/L. Procalcitonin (PCT) was 0.38 ng/ml in the meantime. With tracheal intubation and ventilator-assisted ventilation were given, blood gas values were as follows: pO2, 117 mmHg; pCO2, 31.4 mmHg. After two sets of blood culture were taken, an antibiotic regimen included meropenem (2 g IV, 8 hourly) and vancomycin (1million IU IV 12 hourly) was given. A lung computed tomography (CT) scan was performed on day 3 (Fig. 1) and found patchy consolidation in left inferior lobar. It was considered as traumatic wet lung and/or lung infection. On day 7, blood culture showed no bacteria growth, the hsCRP decreased to 6.7 mg/L, PCT was 0.12 ng/ml, but the white blood cells were still elevated (12.3 × 10E9/L with 89.5% neutrophils). A large amount of Gram-negative rod from the sputum specimen taken at day 5, which identified as Pandoraea Apista by MALDI-TOF MS, was reported at day 8, with no antimicrobial susceptibility test results. The antibiotic regimen remained unchanged because of lack of knowledge about this genus and the infection of this patient seemed to be getting better. However, hsCRP and PCT increased progressively after that. It became more and more difficult to maintain his blood pressure and oxygen saturation. On day 11, the hsCRP was 194.4 mg/L. CT scan (Fig. 2) confirmed new infections in his right lung. The sputum culture result was reconsidered, and the microbiologist of our hospital confirmed that Pandoraea Apista was the only germ grow in the media. After a review of some case report concerning Pandoraea species, meropenem was altered by imipenem (1 g IV, 6 hourly) on day 12. Nonetheless, his condition got worse and the relatives of him asked for a “Do Not Attempt Resuscitation”. He died on day 14 with cardiac respiratory arrest and multiple organ failure.
Discussion and conclusion
The timeline of our case-patient is summarized in Table 1. Pandoraea species is a strange genus to both clinicians and microbiologists in China. It is reasonable to believe that Pandoraea apista might not the responsible pathogen at the time of admission. But the using of broad-spectrum antibiotics promoted the growth of Pandoraea apista, which lead to the right pneumonia and a worsening condition of this patient. To better understand Pandoraea species and its pathogenicity, we present a review from literature reported before November 31, 2018.
Microbiology, distribution, and identification
Pandoraea species belongs to the β-subclass of the Proteobacteria, which contains a group of Gram-negative bacilli that are aerobic or facultative anaerobic (e.g. P. pnomenusa reported by Ambrose et al. [3]), do not form spores, do not reduce nitrate, do not ferment lactose, and rely on flagellar movement. Growth is observed at 30 °C and 37 °C. Catalase activity is variable [4], along with lack of saccharolytic activity, and mostly are o-nitrophenyl-β-D-galactopyranoside (ONPG) negative [1, 2].
It presently comprises five species that have been isolated from human clinical specimens (P. apista, P. pulmonicola, P. pnomenusa, P. sputorum, and P. norimbergensis) [1], five named non-clinical species (P. thiooxydans [5], P. oxalativorans [6], P. faecigallinarum [6], P. vervacti [6], and P. terrae [4]) that have been isolated only from non-clinical origin and at least four unnamed genomospecies [1, 7]. The main sources of Pandoraea species in the environment including soil, animal feces, water, and even powdered milk [8]. The clinical species were mainly isolated from respiratory specimens of cystic fibrosis (CF) patients [1, 3, 7, 9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24]. To date, cases of Pandoraea species caused colonization or infection have been reported all over the world, including USA [1, 7, 12, 25], Denmark [1, 10, 26], Germany [13], France [18, 19], Ireland [26], Argentina [20, 24], Spain [16, 17, 27, 28], Australia [3, 14, 15], Canada [1], UK [1, 9, 22], China [29], Belgium [1], Brazil [1], Sweden [1] (See at Fig. 3). Most cases occurred in Europe, America, and Australia, which was consistent with the epidemiology of CF [30].
From 1974 to 2014, only 34 cases of CF were reported in China [31]. Pandoraea species seem to be rare in non-CF patients, which may be the major reason that the first clinical isolate was reported not until the end of 2018 in China. But in our opinion, the incidence of infections caused by Pandoraea species is underestimated due to the difficulty in identification.
Identification of Pandoraea species through routine diagnostic laboratories, such as phenotypic methods and VITEK 2 automatic microorganisms analyzer, can commonly lead to misidentification [1, 7, 14, 15, 20, 29]. Molecular analysis for further confirmation is necessary when an isolate is unclearly identified. The cellular fatty acid analysis may be useful [1, 2], and yet cellular fatty acid-deficient isolate has been reported [15]. Genus-specific PCR assays and the sequences of 16S rRNA [1, 32] and gyrB [33] genes have proven to be reliable but may have some limitation in differentiating the Pandoraea species [15, 29]. More recently, several studies have reported good results in using MALDI-TOF MS for the identification of Pandoraea species [16,17,18, 20]. MALDI-TOF MS is a quick, easy and practical, high throughput analytic method that relies on a comparison between the mass spectrum of the isolate and the mass spectra in available databases. But Pandoraea species are rare in clinical isolates. With less information contributed by Pandoraea species in the database used might limit the discriminatory power of this method. Misidentification in strains-level has been reported [3]. MALDI-TOF MS is a promising approach, but more specific information is needed to update its database for accurate confirmation of bacteria that is less common. But with MALDI-TOF MS, quick identification, in the beginning, becomes possible. Martina and et al. [24] reported “the first case of Pandoraea sputorum colonization in Argentina” in 2017, but two Pandoraea sputorum strains, along with a Pandoraea apista and a Pandoraea pulmonicola, had been re-identified from 396 non-fermenting Gram-negative bacilli clinical isolates from a hospital in Argentina in 2015 [20]. Colonization and infection associated with Pandoraea species may have always existed, but were missed by the approach we used in the past.
Pathogenicity
The pathogenicity of Pandoraea species in CF patients remains controversial. In many cases, a degree of deterioration has been observed after Pandoraea species were cultured together with some other bacteria in a respiratory specimen. But bacteremia caused by Pandoraea species in CF patients has been reported [11]. Some studies show that Pandoraea species may spread between CF patients [10, 19]. Burkholderia sp., which belongs to the same family as Pandoraea species, is generally considered transmissible and may cause severe infection after lung translation [34]. Many experts strongly recommend isolation for CF patients infected or colonized with Burkholderia cepacia [35]. Although the study of Pimentel et al. [14] shows that colonized with Pandoraea species before lung transplantation in CF patients may not be a predictor of poor outcomes after transplantation. It is important to find out the pathogenicity and transmissibility of Pandoraea sp. in CF patients.
Reports of non-CF patients infected or colonized with Pandoraea species are summarized in Table 2. Unlike CF patients, Pandoraea infection seemed to be more likely to cause bacteremia in non-CF patients [1, 7, 25, 27, 29], and co-presenting with other pathogens is only reported in two cases [25, 28]. In the study of Coenye et al. [1] and Daneshvar et al. [7], Pandoraea species cultured from respiratory specimens of non-CF patients have been reported, but with less case information. It makes our case the first well-reported case of pneumonia potentially caused by Pandoraea species in non-CF patients. Pneumonia was also reported in the case of Stryjewski et al. [25]. And yet, Pandoraea pnomenusa was cultured only from his blood samples, but not from respiratory samples. Furthermore, co-existing with nocardiosis and mycetomas makes it confusing to identify the responsible pathogen of pneumonia. It is reasonable to see Pandoraea species as an opportunistic pathogen in non-CF patients.
Some studies have investigated the pathogenesis of Pandoraea species. The ability to trigger a pronounced pro-inflammatory response, with an elevation of both interleukin (IL)-6 and IL-8 has been reported in the study of Caraher et al. [26]. This study also demonstrates that only a few strains have the abilities to invade lung epithelial cells (3 out of 19) and form biofilms (1 out of 19). According to Costello et al. [36], cellular invasion of Pandoraea species is independent of CF phenotype, and Pandoraea strains were also capable of translocation across polarized lung epithelial cell monolayers. The lack of enhanced susceptibility to the invasion of cells with a CF phenotype over non-CF cells is also discovered in their study. This may be one of the reasons that Pandoraea species more commonly lead to colonization rather than bacteremia in CF patients. Further research [37] shows that co-colonized with Pseudomonas aeruginosa may be another reason that Pandoraea species behave more gently in CF patients. Pseudomonas aeruginosa can inhibit the growth of P. pulmonicola and P. apista and the pro-inflammatory effects caused by these strains. These findings agree with a summary of Pandoraea species infections in transplant patients produced by Pimentel et al. [14], in which 5 cases of lung transplant patients have been reviewed and found out that CF patients previously colonized with Pandoraea species and Pseudomonas aeruginosa seem to have a better survival after transplantation than the only one non-CF patient, who was without Pseudomonas aeruginosa colonized previously and died after transplantation due to Pandoraea pnomenusa bacteremia.
Molecular biotechnology has been used to analyze virulence genes and drug resistance genes of Pandoraea species. Lim et al. [22] sequenced the complete genome of a Pandoraea pnomenusa strain and identified 16 virulence factors, which are well-characterized virulence determinants in some other pathogens. Robson et al. [38] sequenced the complete genome of two Pandoraea pnomenusa strains and found 130 gene sequences related to virulence, disease and drug resistance.
Mobile genetic elements (MGE), such as plasmids, can spread virulence and antibiotic resistance genes between microbes [39]. Yong et al. [21] analyzed one plasmid from Pandoraea apista and 7 plasmids from non-clinical Pandoraea strains (Pandoraea faecigallinarum, Pandoraea thiooxydans, and Pandoraea vervacti). More virulence genes were found in non-clinical strains than in Pandoraea apista and antibiotic resistance genes were only detected in the plasmids from non-clinical Pandoraea strains. This means that the Pandoraea apista strain analyzed is less virulent and lack of the ability to spread antibiotic resistance genes through plasmid. But with only one clinical strain detected, we do not know if this conclusion could be extended to all the clinical Pandoraea strains. However, many opportunistic pathogens are found transmit into clinical settings from the environment nowadays. These non-clinical strains may also be a thread.
Susceptibility
In the study of Daneshvar et al. [7] in 2001, susceptibility result of some strains is not given individually. So we take the isolates from the same strain of Pandoraea as one isolate and using the mode minimum inhibitory concentration (MIC) to determine the susceptibility.
To dates, there are no breakpoints for the results of antimicrobial susceptibility tests suggested for Pandoraea species. Interpretive susceptibility criteria suggested for Burkholderia cepacia complex [19], other Non-Enterobacteriaceae [12, 17, 24], Pseudomonas aeruginosa [3, 29], or Stenotrophomonas maltophilia [3] was used to determine the results of susceptibility tests. Different criteria, and in some cases, different susceptibility methods might lead to unavoidable bias. Antimicrobial susceptibility profiles of Pandoraea species reported in the literature are summarized in Table 3. From Table 3, we can tell that Pandoraea is resistant to most antibiotic agents in most cases, such as aminoglycosides, most β-lactam agents and quinolones. However, the sensitivity of Pandoraea to piperacillin, piperacillin-tazobactam, aminoglycosides, and fluoroquinolones is variable. Pandoraea sputorum seems to be more sensitive to piperacillin-tazobactam than any other strains. Pandoraea apista is the only strain that shows sensitivity to ciprofloxacin and sparfloxacin in some studies. In contrast, most Pandoraea strains are sensitive to imipenem, tetracycline, and trimethoprim-sulfamethoxazole. It is important to note that almost all the Pandoraea isolates demonstrated resistance to meropenem but most of the strains are sensitive to imipenem, which is exactly on the opposite of Burkholderia. Even though they are closely related. Agents that have a potential activity to Pandoraea genus but fewer data reported include doxycycline [11], minocycline [27], tigecycline [18], and rifampicin [18, 19].
Enzyme-production is one of the most important mechanisms of bacterial resistance to antibacterial agents. Carbapenems are essential for some severe multi-drug resistant bacterial infections. Germs that can produce carbapenemases are seeing as a great threat to human beings [41]. Schneider et al. [13] found out that Oxacillinases-62 (OXA-62) is involved in the mechanism of resistance to imipenem. OXA-62 is only reported in P. pnomenusa species, and yet resistance to imipenem of Pandoraea apista [12] and Pandoraea pulmonicola [18, 19] have been reported, indicating that there may be more than one mechanism involved. The MICs of meropenem in this study was reduced by 8 times after adding efflux pump inhibitors, which indicating resistance of P. pnomenusa to meropenem may contribute by two mechanisms including producing OXA as well as an efflux pump. The researchers subsequently sequenced the oxacillinases of nine isolates belong to six Pandoraea species and found nine novel oxacillinase variants (OXA151-OXA159) [40]. 1All the strains are resistant to meropenem, but the MICs of meropenem can be reduced by 4–32 times by adding an active-site serine β-lactamases inhibitor, confirmed that these new oxacillinases also have the ability to hydrolyze meropenem.
Pandoraea. is a new genus that has been classified within 20 years. We report a case of a patient admitted to the ICU after removal of traumatic intracranial hematoma and decompressive craniectomy, who subsequently developed Pandoraea related pneumonia and eventually died of multiple organ failure. Through a literature review, we learned that Pandoraea sp. is a multi-drug resistant opportunistic pathogen, which can cause pneumonia and bacteremia by several mechanisms. Although this bacterium is more commonly found in CF patients, there have been reports of infection in non-CF patients, and there is evidence supporting Pandoraea species could be more virulent in non-CF patients. The genus is usually sensitive to imipenem, tetracycline, and SMT. However, the susceptibility is highly variable. Species-level identification and antibiotic susceptibility test are necessary.
Availability of data and materials
All the data and material involved in the current study are available from the corresponding author on reasonable request.
Abbreviations
- CF:
-
Cystic fibrosis
- COPD:
-
Chronic obstructive pulmonary disease
- CT:
-
Computed tomography
- EICU:
-
Emergency intensive care unit
- hsCRP:
-
hypersensitive C-reactive protein
- IL:
-
Interleukin
- MALDI-TOF MS:
-
Matrix-assisted laser desorption ionization–time-of-flight mass spectrometry
- MIC:
-
Minimum inhibitory concentration
- OXA:
-
Oxacillinases
- PCT:
-
Procalcitonin
- PFGE:
-
Pulsed field gel electrophoresis
References
Coenye T, Falsen E, Hoste B, Ohlén M, Goris J, Govan JRW, Gillis M, Vandamme P. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int J Syst Evol Micr. 2000;50 Pt 2(2):887. https://doi.org/10.1099/00207713-50-2-887.
Lipuma JL, Currie BJ, Peacock SJ, et al. Burkholderia, Stenotrophomonas, Ralstonia, Cupriavidus, Pandoraea, Brevundimonas, Comamonas, Delftia, and Acidovorax. In: Jorgensen JH, editor. Manual of clinical microbiology. 11th ed. Washington DC: American Society of Microbiology Press; 2015. p. 791–803. https://doi.org/10.1128/9781555817381.
Ambrose M, Malley RC, Warren SJ, Beggs SA, Swallow OF, McEwan B, Stock D, Roddam LF. Pandoraea pnomenusa isolated from an Australian patient with cystic fibrosis. Front Microbiol. 2016;7:692. https://doi.org/10.3389/fmicb.2016.00692.
Jeong SE, Lee HJ, Jia B, Jeon CO. Pandoraea terrae sp. nov., isolated from forest soil and emended description of the genus Pandoraea Coenye et al., 2000. Int J Syst Evol Micr. 2016, 66(9). https://doi.org/10.1099/ijsem.0.001229.
Anandham R, Indiragandhi P, Kwon SW, Sa TM, Jeon CO, Kim YK, Jee HJ. Pandoraea thiooxydans sp. nov., a facultatively chemolithotrophic, thiosulfate-oxidizing bacterium isolated from rhizosphere soils of sesame (Sesamum indicum L.). Int J Syst Evol Micr. 2010;60(Pt 1):21. https://doi.org/10.1099/ijs.0.012823-0.
Sahin N, Tani A, Kotan R, Sedláček I, Kimbara K, Tamer AU. Pandoraea oxalativorans sp. nov., Pandoraea faecigallinarum sp. nov. and Pandoraea vervacti sp. nov., isolated from oxalate-enriched culture. Int J Syst Evol Micr. 2010;61(Pt 9). https://doi.org/10.1099/ijs.0.026138-0.
Daneshvar MI, Hollis DG, Steigerwalt AG, Whitney AM, Spangler L, Douglas MP, Jordan JG, MacGregor JP, Hill BC, Tenover FC, et al. Assignment of CDC weak oxidizer group 2 (WO-2) to the genus Pandoraea and characterization of three new Pandoraea genomospecies. J Clin Microbiol. 2001;39(5):1819–26. https://doi.org/10.1128/JCM.39.5.1819-1826.2001.
Moore JE, Coenye T, Vandamme P, Stuart Elborn J. First report of Pandoraea norimbergensis isolated from food—potential clinical significance. Food Microbiol. 2001;18(1):113–4. https://doi.org/10.1006/fmic.2000.0375.
Moore JE, Reid A, Millar BC, Jiru X, Mccaughan J, Goldsmith CE, Collins J, Murphy PG, Elborn JS. Pandoraea apista isolated from a patient with cystic fibrosis: problems associated with laboratory identification. Brit J Biomed Sci. 2002;59(3):164–6. https://doi.org/10.1080/09674845.2002.11978036.
Jørgensen IM, Johansen HK, Frederiksen B, Pressler T, Hansen A, Vandamme P, Høiby N, Koch C. Epidemic spread of Pandoraea apista, a new pathogen causing severe lung disease in cystic fibrosis patients. Pediatr Pulm. 2003;36(5):439–46. https://doi.org/10.1002/ppul.10383.
Johnson LN, Han JY, Moskowitz SM, Burns JL, Qin X, Englund JA. Pandoraea bacteremia in a cystic fibrosis patient with associated systemic illness. Pediatr Infect Dis J. 2004;23(9):881–2. https://doi.org/10.1097/01.inf.0000136857.74561.3c.
Atkinson RM, Lipuma JJ, Rosenbluth DB, Dunne WM. Chronic colonization with Pandoraea apista in cystic fibrosis patients determined by repetitive-element-sequence PCR. J Clin Microbiol. 2006;44(3):833–6. https://doi.org/10.1128/JCM.44.3.833-836.2006.
Schneider I, Queenan AM, Bauernfeind A. Novel carbapenem-hydrolyzing oxacillinase OXA-62 from Pandoraea pnomenusa. Antimicrob Agents Ch. 2006;50(4):1330–5. https://doi.org/10.1128/AAC.50.4.1330-1335.2006.
Pimentel JD, MacLeod C. Misidentification of Pandoraea sputorum isolated from sputum of a patient with cystic fibrosis and review of Pandoraea species infections in transplant patients. J Clin Microbiol. 2008;46(9):3165–8. https://doi.org/10.1128/JCM.00855-08.
Aravena-Román M. Cellular fatty acid-deficient Pandoraea isolated from a patient with cystic fibrosis. J Med Microbiol. 2008;57(Pt 2):252. https://doi.org/10.1099/jmm.0.47671-0.
Martínez-Lamas L, Rabade Castedo C, Martín Romero Domínguez M, Barbeito Castiñeiras G, Palacios Bartolomé A, Pérez del Molino Bernal ML. Pandoraea sputorum colonization in a patient with cystic fibrosis. Arch Bronconeumol. 2011;47(11):571–4. https://doi.org/10.1016/j.arbr.2011.06.014.
Fernández-Olmos A, Morosini MI, Lamas A, García-Castillo M, García-García L, Cantón R, Máiz L. Clinical and microbiological features of a cystic fibrosis patient chronically colonized with Pandoraea sputorum identified by combining 16S rRNA sequencing and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50(3):1096–8. https://doi.org/10.1128/JCM.05730-11.
Kokcha S, Bittar F, Reynaud-Gaubert M, Mely L, Gomez C, Gaubert JY, Thomas P, Rolain JM. Pandoraea pulmonicola chronic colonization in a cystic fibrosis patient, France. New Microbes New Infect. 2013;1(2):27–9. https://doi.org/10.1002/2052-2975.16.
Degand N, Lotte R, Decondé Le Butor C, Segonds C, Thouverez M, Ferroni A, Vallier C, Mély L, Carrère J. Epidemic spread of Pandoraea pulmonicola in a cystic fibrosis center. BMC Infect Dis. 2015;15:583. https://doi.org/10.1186/s12879-015-1327-8.
Almuzara M, Barberis C, Traglia G, Famiglietti A, Ramirez MS, Vay C. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry for species identification of nonfermenting gram-negative bacilli. J Microbiol Meth. 2015;112:24–7. https://doi.org/10.1016/j.mimet.2015.03.004.
Yong D, Tee KK, Yin WF, Chan KG. Characterization and comparative overview of complete sequences of the first plasmids of Pandoraea across clinical and non-clinical strains. Front Microbiol. 2016;7:1606. https://doi.org/10.3389/fmicb.2016.01606.
Lim YL, Ee R, Yong D, Yu CY, Ang GY, Tee KK, Yin WF, Chan KG. Complete genome sequence analysis of Pandoraea pnomenusa type strain DSM 16536(T) isolated from a cystic fibrosis patient. Front Microbiol. 2016;7:109. https://doi.org/10.3389/fmicb.2016.00109.
Dupont C, Aujoulat F, Chiron R, Condom P, Jumas-Bilak E, Marchandin H. Highly diversified Pandoraea pulmonicola population during chronic colonization in cystic fibrosis. Front Microbiol. 2017;8:1892. https://doi.org/10.3389/fmicb.2017.01892.
Martina PF, Martínez M, Frada G, Alvarez F, Leguizamón L, Prieto C, Barrias C, Bettiol M, Lagares A, Bosch A, et al. First time identification of Pandoraea sputorum from a patient with cystic fibrosis in Argentina: a case report. BMC Pulm Med. 2017;17(1):33. https://doi.org/10.1186/s12890-017-0373-y.
Stryjewski ME, LiPuma JJ, Messier RH, Reller LB, Alexander BD. Sepsis, multiple organ failure, and death due to Pandoraea pnomenusa infection after lung transplantation. J Clin Microbiol. 2003;41(5):2255–7. https://doi.org/10.1128/jcm.41.5.2255-2257.2003.
Caraher E, Collins J, Herbert G, Murphy PG, Gallagher CG, Crowe MJ, Callaghan M, McClean S. Evaluation of in vitro virulence characteristics of the genus Pandoraea in lung epithelial cells. J Med Microbiol. 2008;57(Pt 1):15–20. https://doi.org/10.1099/jmm.0.47544-0.
Falces-Romero I, Gutiérrez-Arroyo A, Romero-Gómez MP. Catheter-associated bacteremia by Pandoraea pnomenusa in an infant with acute lymphoblastic leukemia. Med Clin-Barcelona. 2016;147(3):132. https://doi.org/10.1016/j.medcli.2016.04.009.
Monzón T, Valga F, Reichert J, López C. Hemodialysis catheter colonized by Pandoraea spotorum. Nefrologia. 2018;38(6):662–4. https://doi.org/10.1016/j.nefro.2017.11.003.
Gao J, Wei Q, Shao HC, Wu H. A case report of Pandoraea sp. isolated from neonatal blood culture. Chin J Microbes Infec. 2018;13(06):368–72 (in Chinese with English abstract).
Elborn JS. Cystic fibrosis. Lancet. 2016;388(10059):2519–31. https://doi.org/10.1016/S0140-6736(16)00576-6.
Shen Y, Liu J, Zhong L, Mogayzel PJ, Zeitlin PL, Sosnay PR, Zhao S. Clinical phenotypes and genotypic Spectrum of cystic fibrosis in Chinese children. J Pediatr. 2016;171:269–76. https://doi.org/10.1016/j.jpeds.2015.12.025.
Coenye T, Liu L, Vandamme P, LiPuma JJ. Identification of Pandoraea species by 16S ribosomal DNA-based PCR assays. J Clin Microbiol. 2001;39(12):4452–5. https://doi.org/10.1128/JCM.39.12.4452-4455.2001.
Coenye T, Lipuma JJ. Use of the gyrB gene for the identification of Pandoraea species. FEMS Microbiol Lett. 2002. https://doi.org/10.1016/S0378-1097(01)00589-4.
Biddick R, Spilker T, Martin A, Lipuma JJ. Evidence of transmission of Burkholderia cepacia , Burkholderia multivorans and Burkholderia dolosa among persons with cystic fibrosis. FEMS Microbiol Lett. 2010;228(1):57–62. https://doi.org/10.1016/S0378-1097(03)00724-9.
Vonberg RP, Gastmeier P. Isolation of infectious cystic fibrosis patients: results of a systematic review. Infect Control Hosp Epidemiol. 2005;26(4):401–9. https://doi.org/10.1086/502558.
Costello A, Herbert G, Fabunmi L, Schaffer K, Kavanagh KA, Caraher EM, Callaghan M, McClean S. Virulence of an emerging respiratory pathogen, genus Pandoraea, in vivo and its interactions with lung epithelial cells. J Med Microbiol. 2011;60(Pt 3):289–99. https://doi.org/10.1099/jmm.0.022657-0.
Costello A, Reen FJ, O'Gara F, Callaghan M, McClean S. Inhibition of co-colonizing cystic fibrosis-associated pathogens by Pseudomonas aeruginosa and Burkholderia multivorans. Microbiology. 2014;160(Pt 7):1474–87. https://doi.org/10.1099/mic.0.074203-0.
Ee R, Ambrose M, Lazenby J, Williams P, Chan KG, Roddam L. Genome Sequences of Two Pandoraea pnomenusa Isolates Recovered 11 Months Apart from a Cystic Fibrosis Patient. Genome Announc. 2015;3(1). https://doi.org/10.1128/genomeA.01389-14.
von Wintersdorff CJ, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, Savelkoul PH, Wolffs PF. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol. 2016;7:173. https://doi.org/10.3389/fmicb.2016.00173.
Schneider I, Bauernfeind A. Intrinsic carbapenem-hydrolyzing oxacillinases from members of the genus Pandoraea. Antimicrob Agents Chemother. 2015;59(11):7136–41. https://doi.org/10.1128/AAC.01112-15.
Wang Q, Wang X, Wang J, Ouyang P, Jin C, Wang R, Zhang Y, Jin L, Chen H, Wang Z, et al. Phenotypic and Genotypic Characterization of Carbapenem-resistant Enterobacteriaceae: Data From a Longitudinal Large-scale CRE Study in China (2012–2016). Clin Infect Dis. 2018;67(suppl_2):S196–205. https://doi.org/10.1093/cid/ciy660.
Acknowledgments
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
CL and PY were responsible for the conception and design of the study. CL, NL, JZ, and MC were responsible for acquisition of data. CL, NL, and PY drafted the manuscript. PY, NL, QX, and GZ revised and commented on the draft. All authors read and approved the final version of the manuscript. None of the authors have any competing interests.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Written informed consent was obtained from the patient’s family members for publication of this case report and any accompanying images. A copy of the written consent is available for review by the editor of this journal.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lin, C., Luo, N., Xu, Q. et al. Pneumonia due to Pandoraea Apista after evacuation of traumatic intracranial hematomas:a case report and literature review. BMC Infect Dis 19, 869 (2019). https://doi.org/10.1186/s12879-019-4420-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-019-4420-6