Abstract
Background
Endograft infection is a rare but extremely dangerous complication of aortic repair (25–100% of mortality). We describe here the first case of Listeria monocytogenes abdominal periaortitis associated with a vascular graft. We also discuss the differential diagnosis of periaortitis and provide a literature review of L. monocytogenes infectious aortitis.
Case presentation
Nine months after endovascular treatment of an abdominal aortic aneurysm (abdominal stent graft), a 76-year-old man was admitted for severe abdominal pain radiating to the back. Laboratory tests were normal apart from elevated C-reactive protein (CRP). Injected abdominal computed tomography (CT) showed infiltration of the fat tissues around the aortic endoprosthesis and aneurysmal sac expansion; positron emission tomography with 2-deoxy-2-[fluorine-18]fluoro- D-glucose integrated with computed tomography (18F-FDG PET/CT) showed a hypermetabolic mass in contact with the endoprosthesis. Blood cultures were negative. At surgical revision, an infra-renal peri-aortic abscess was evident; post-operative antibiotic therapy with ciprofloxacin and doxycycline was started. Cultures of intraoperative samples were positive for L. monocytogenes. Results were further confirmed by a broad-range polymerase chain reaction (PCR) and next-generation sequencing. Antibiotic treatment was switched to intravenous amoxicillin for 6 weeks. Evolution was uneventful with decrease of inflammatory parameters and regression of the abscess.
Conclusion
An etiologic bacterial diagnosis before starting antibiotic therapy is paramount; nevertheless, culture-independent methods may provide a microbiological diagnosis in those cases where antimicrobials are empirically used and when cultures remain negative.
Similar content being viewed by others
Background
Aortic inflammatory disease is rare (1.38 cases/100,000 inhabitants) and prevails in middle aged (50–60 years old) men (male/female ratio 2:1-3:1). It comprises two main entities: aortitis and periaortitis, both characterised by the infiltration of the arterial layer by inflammatory cells and the disruption of the arterial structure [1]. Nevertheless, while aortitis is confined to the arterial wall, in (chronic) periaortitis adventitial inflammation develops and leads to the involvement of adjacent retroperitoneal structures (e.g. ureters and inferior vena cava). Thoracic involvement has been described, despite the fact that this infection typically affects the infrarenal aorta and/or the iliac arteries.
Chronic periaortitis is classically considered to encompass three distinct entities: idiopathic retroperitoneal fibrosis, inflammatory abdominal aortic aneurysms and perianeurysmal retroperitoneal fibrosis [2]. Clinically indistinguishable (i.e. dull abdominal pain, fever, fatigue, weight loss) and histologically similar (i.e. a thick fibro-inflammatory infiltrate rich in lymphocytes and collagen), some authors hypothesize a spectrum of the same disease [3]. Once considered a local complication of aortic atherosclerosis, chronic periaortitis seems rather to be a manifestation of a systemic disease (e.g. ankylosing spondylitis, rheumatoid arthritis, IgG4-related disease, Wegener’s granulomatosis, polyarteritis nodosa) [4]. If an underlying immune disease is excluded, the clinician should actively investigate for more unusual causes, like tumors, drugs, radiotherapy, trauma, major abdominal surgery, myeloproliferative disorders (Erdheim-Chester disease), exposure to toxics (e.g. asbestos) and drugs (e.g. ergot-derived drugs) as well as infections [5].
Before the beginning of the “antibiotic era”, the frequency of aortic infections ranged from 2.6 to 3.4% [6]. Although aortic infections are nowadays much rarer, they remain life-threatening. Antibiotics, indeed, lead to the near complete disappearance of tuberculous and syphilitic aortitis, as well as a drastic reduction in endocarditis-associated septic embolization and mycotic aneurysms (“mycotic” is misnomer: Osler used this term to describe the “fresh fungous” appearance of these bacterial lesions) [7].
Until the intimal layer is intact, the aorta is extremely resistant to infections. Therefore, haematogenous infections often arise in the context of a damaged endothelium, like in atherosclerotic lesions, aneurysms, aortitis, and congenital abnormalities. Besides bacteraemia, which is the most likely route of infection, septic embolization to the vasa vasorum (as in infectious endocarditis), extension form a neighbouring infectious focus as well as contamination in the setting of intravenous drug use, trauma and invasive intravascular procedures have been described [8].
Infectious periaortitis seems to occur even less frequently than aortitis, [9] with only one possible case appearing in medical literature [10]. We believe that insufficient or unreliable diagnostic techniques are likely responsible for an underestimation of this condition.
From the infectious diseases standpoint, clinicians should consider the possibility of infections sustained by fastidious organisms commonly missed by conventional cultures, e.g. Brucella spp. and L. monocytogenes.
In this paper, we describe the first reported case of L. monocytogenes abdominal periaortitis. Moreover, we provide a review of the literature and we discuss about a culture-independent method for microbiological diagnosis.
Case presentation
A 76-year-old man with a history of chronic obstructive pulmonary disease (Gold grade 4) and type 2 diabetes was admitted for abdominal pain radiating to the back for 10 days; pain was punctuated by brief episodes of diarrhoea. Nine months before admission, the patient had undergone endovascular treatment of an abdominal aortic aneurysm (abdominal stent graft).
Patient was afebrile, and physical examination was unremarkable apart from diffuse abdominal tenderness without guarding or rebound. Laboratory tests showed mild anaemia (111 g/L), a total white blood cell count of 7.6 × 109/l with a neutrophil count of 5.21 × 109/l; CRP was 233.5 mg/l and procalcitonin 0.09 μg/L. Kidney (creatinine, urea, sodium and potassium dosage) and liver (ALT, SGOT, SGPT, bilirubin, gamma-glutamyltranspeptidase dosage) function tests were within normal ranges.
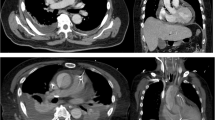
A contrast-enhanced abdominal computed tomography (CT) scan showed infiltration of the fatty tissues around the aortic endoprosthesis and increase of the aneurysmal sac expansion by 6.0 mm (57 versus 51 mm) compared to a previous CT performed 2 months earlier; no endoleak was observed. No other abnormalities were observed. 18F-FDG PET/CT (Fig. 1, a-e) showed a hypermetabolic (SUVmax = 8.5) mass with a diameter of 15 mm in contact with the superior antero-medial of the endoprosthesis, suggesting an abscess. CT colonography excluded neoplasia.
Serum immunofixation was normal, without increase in the IgG4 subclass. The interferon-gamma reactivitiy assay was negative. Serologies for Treponema pallidum, Bartonella henselae and Coxiella burnetii were negative, as were Brucella agglutination tests (Rose-Bengal and Wright). Multiple blood cultures showed no growth. Real-Time Quantitative broad-range PCR on blood was negative. Stool PCR testing for Campylobacter jejuni/coli, Shigella spp. and Salmonella spp. was negative.
Multidisciplinary consensus involving interventional radiologists, vascular surgeons, infectious disease specialists, and internists, led to the decision for a surgical revision. During the procedure, an infrarenal peri-aortic abscess was noted, from which samples were performed. In the post-operative period and before culture results were available, patient was started on oral ciprofloxacin (500 mg/12 h) and doxycyclin (100 mg/12 h). Histopathological examination of the intraoperative specimens revealed fibrous tissue with accompanying inflammation rich in histiocytes and cholesterol crystals; no microorganisms were identified on hematoxylin/eosin, Gram or Grocott stains.
Cultures of intraoperative samples (5/5) became positive for L. monocytogenes after 10 days incubation. These results were further confirmed by a broad-range PCR. Susceptibility testing (Kirby-Bauer disk diffusion susceptibility tests (SirScan 200 Automatic, i2A, France) interpreted according to the EUCAST clinical MIC breakpoints) [11] showed a pan-susceptible (“wild type”) L. monocytogenes strain with a minimum inhibitory concentration [MIC] for amoxicillin of 0.380 mg/l. Antibiotic treatment was then switched to intravenous amoxicillin ((2 g/6 h) for a duration of 6 weeks. The patient’s clinical course was favourable, and CRP levels became within normal range. At the end of antibiotic course, an abdominal CT scan showed regression of the peri-aortic abscess and collection. At 6 months follow-up the patient was well, without recurrence.
DNA extraction
Frozen aortic tissue was cut into small pieces on a disposable Petri dish support using a scalpel. DNA was extracted from 83 mg of shredded sample using the Ultra-Deep Microbiome Prep kit (Molzym, Bremen, Germany) for enrichment of bacteria/fungal DNA, according to the manufacturer’s instructions (Version 2.0) for tissue samples.
Quantitative PCR (qPCR) assays
The concentration of bacterial and human DNA was determined by qPCR experiments as previously described [12], using 16S rRNA and beta-actin reference genes, respectively. The reference curves for bacterial and human DNA quantitation were generated using known concentrations of Escherichia coli DH5α genomic DNA and human genomic DNA from the TaqMan beta-Actin Detection Reagent kit (Applied Biosystems, Framingham, MA), respectively.
DNA sequencing
Metagenomic libraries were prepared from 1 ng DNA (sum of bacterial and human DNA load determined by qPCR), using Nextera XT DNA Sample Preparation Kit according to Illumina (San Diego, USA) instructions, except that 16 (instead of 12) PCR enrichment cycles were used. The libraries were sequenced for 2 × 250 + 8 cycles on an Illumina MiSeq instrument at Fasteris (Plan-les-Ouates, Switzerland) using the MiSeq Reagent Kit v3 and MiSeq Control Software 2.6.2.1.
Bioinformatics analysis
The Trimmomatic package was used by the sequencing service provider to remove bases that correspond to the standard Illumina adapters and to trim low-quality ends of reads at the beginning of a 4-base wide sliding window with an average Phred quality < 5 [13]. We further trimmed the reads with the low-quality base score of Q10 and a mean quality score Q30 over a sliding 20-base window. Any read that, after trimming, had a length < 150 bases was discarded. To filter out putative artificial replicate reads, we used a home-made script which retains only the longest sequence from those with identical first 100 bases in either forward or reverse reads. Unpaired forward or reverse reads were discarded. Remaining reads were classified with Kraken v.0.10.5-beta with the default parameters [14]. After filtering out the read pairs that matched human genome, the data were deposited to European Nucleotide Archive (ENA) database (accession number PRJEB21816).
The reads classified by Kraken to genus Listeria were retrieved and mapped to complete reference genomic sequences of Listeria strains (downloaded from the NCBI Reference Sequence (RefSeq) prokaryotic genome collection) [15] using USEARCH 8.1.11861 [16] (−ublast -id 0.98 -cov 0.98 -top_hits_only -strand both -evalue 0.00001). The number of hits was counted for each Listeria strain. The strain with the maximum number of hits was retained as a reference for read mapping using GView [17]. MetaPhlAn2 taxonomic profiling, based on read mapping against ~ 1 million markers from more than 7500 species was used with default settings [18] (Fig. 2).
Number and taxonomic assignments of sequence reads obtained by WMGS of DNA extracts from the perioaortic tissue sample. F, forwards reads; R, reverse reads. a. Number of sequence reads at different steps of data analysis. b. Krona plots of bacterial taxa identified by Kraken [34]. c. Ring diagram showing the genome of the L. monocytogenes strain 6179 chromosome sequence (GenBank accession number NZ_HG813249). Outer circle depicts the chromosome regions to which mapped pairs of sequence reads indicated by bars. Middle circle shows G + C content expressed as percentage. The inner ring indicates chromosomal position. The figure was generated using GView [17]
Although culture and broad-range PCR detected L. monocytogenes in the clinical specimen, we performed next-generation sequencing in order to determine the clinical utility of such a technique. We extracted DNA from the aortic tissue using a protocol that improves the bacterial-to-human DNA ratio based on selective lysis of host cells and removal of host DNA before the lysis of bacterial cells [19, 20]. The concentrations of human and bacterial DNA in purified extracts were 5983 and 214 pg/μL, respectively, as determined by qPCR assays. The whole metagenome shotgun sequencing generated 5,509,950 raw paired reads of which 3,026,609 passed quality control (Fig. 1a). In line with qPCR results, most reads (97.7%) corresponded to human genome sequence. Of 66,451 read pairs classified by Kraken as bacterial, 65,805 (99%) were assigned to the genus Listeria and 64,035 (96.4%) to L. monocytogenes (Fig. 1 b, c). Most of the remaining bacterial reads corresponded to Propionibacterium (464 reads) and Micrococcus (32 reads), both of which are frequent reagent contaminants. We have previously identified Propionibacterium in negative extraction controls (PRJEB20595) obtained with a DNA extraction kit of the same lot [20]. Mapping of sequencing reads against clade-specific marker genes using MetaPhlAn2 confirmed the results obtained by Kraken. The MetaPhlAn2 taxonomic profiling showed high abundance of Listeria (99.91%) and L. monocytogenes (96.28%), with a minor presence of Cutibacterium acnes (formerly Propionibacterium acnes) (0.089%).
Discussion and conclusions
Endograft infection after aortic repair can spread to the arterial wall and disrupt it, causing graft migration, aortic dilatation and rupture. This condition is rare, occurring between < 1 and 3% following endovascular or open procedures to the thoracic aorta, and between 0.4 and 1.3% following endovascular or open abdominal aortic repair. Mortality is high 25–100%, with higher rates when a conservative strategy is adopted [21, 22].
Whereas Staphylococci and Salmonella spp. (in particular Salmonella enterica serovar Typhimurium, Enteritidis and Cholerasuis) are the most common microorganisms isolated in native aorta infections, accounting for approximately 50–60% and 30–40% of all cases respectively, Staphylococci, E.coli and Pseudomonas aeruginosa predominate in endograft infections [23]. Clinicians should consider the possibility of fastidious organisms commonly missed by conventional cultures (e.g. Brucella spp. and L. monocytogenes). Bona fide mycotic aortitis and endograft infection have been rarely reported [24].
To date only 18 cases of infectious aortitis due to L. monocytogenes have been described. Of these, 2 occurred in the context of thoracic aortic aneurysms [25, 26], 9 in abdominal aortic aneurysms [27,28,29] and 7 in endovascular aortic graft [30,31,32,33]. Our case is, to the best of our knowledge, the first case of L. monocytogenes abdominal periaortitis associated with a vascular graft.
In this case, diagnosis was made only through surgical sampling (blood cultures were all negative, despite absence of any antimicrobial therapy). We strongly emphasize the need to establish an etiologic bacterial diagnosis by avoiding empiric and precipitated use of antimicrobials, as evidenced here. Unbiased, culture-independent methods could help elucidating cases where antimicrobials are empirically used, nevertheless these methods are often non-affordable nor fully evaluated yet. Standard, state-of-the-art diagnostic methods are still based on bacterial culture (on conventional culture media) performed before initiation of antibiotics.
Given this quite unexpected result and the high risk of false-negative culture results, if antibiotics had been administered prior to surgery, we wonder whether an unbiased, culture-independent method could have provided this microbiological diagnosis.
Abbreviations
- 18F-FDG PET/CT:
-
Positron emission tomography with 2-deoxy-2-[fluorine-18]fluoro- D-glucose integrated with computed tomography
- CRP:
-
C-reactive protein
- CT:
-
Computed tomography
- MIC:
-
Minimum inhibitory concentration
- PCR:
-
Polymerase chain reaction
- qPCR:
-
Quantitative polymerase chain reaction
References
Vaglio A, Salvarani C, Buzio C. Retroperitoneal fibrosis. Lancet. 2006;367:241–51.
Töpel I, Zorger N, Steinbauer M. Entzündliche Erkrankungen der Aorta. Gefässchirurgie. 2016;21:80–6.
Chang-Hee S. Chronic Periaortitis as a systemic autoimmune disease, autoimmune disorders, Clio P. Mavragani, IntechOpen, DOI: https://doi.org/10.5772/19274. Available from: https://www.intechopen.com/books/autoimmune-disorders-pathogenetic-aspects/chronic-periaortitis-as-a-systemic-autoimmune-disease
Ramshaw AL, Parums DV. The distribution of adhesion molecules in chronic periaortitis. Histopathology. 1994;24:23–32.
Urban ML, Palmisano A, Nicastro M, Corradi D, Buzio C, Vaglio A. Idiopathic and secondary forms of retroperitoneal fibrosis: a diagnostic approach. Revue Med Interne. 2015;36:15–21.
Miller DV, Oderich GS, Aubry M-C, Panneton JM, Edwards WD. Surgical pathology of infected aneurysms of the descending thoracic and abdominal aorta: Clinicopathologic correlations in 29 cases (1976 to 1999). Hum Pathol. 2004;35:1112–20.
Osler W. The Gulstonian lectures, on malignant endocarditis. British Med J. 1885;1:577–9.
Fowler VG Jr, Scheld WM, Bayer AS. Endocarditis and intravascular infections. In: Mandell GL, Bennett JE, Dolin R, eds. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 7th ed. Philadelphia, PA: Churchill Livingstone/Elsevier, 2010:1067–1112.
Ladich E, Yahagi K, Romero ME, et al. Vascular diseases: aortitis, aortic aneurysms, and vascular calcification. Cardiovasc Pathol. 2016;25:432–41.
Pizzolato E, Bernardi E, Tosello F, et al. A “mushroom” grown up from the aorta. ITJEM-Simeu [Accessed 2 Apr 2018] 2017. http://www.itjem.org/articoli-scientifici/brief-report-and-case-report/314-elisa-pizzolato
Bauer AW, Kirby W, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–6.
Lazarevic V, Gaïa N, Emonet S, Girard M, Renzi G, Despres L, Wozniak H, Yugueros Marcos J, Veyrieras JB, Chatellier S, van Belkum A, Pugin J, Schrenzel J. Challenges in the culture-independent analysis of oral and respiratory samples from intubated patients. Front Cell Infect Microbiol. 2014;4:65.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Davis MPA, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods. 2013;63:41–9.
Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1.
Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G. Interactive microbial genome visualization with GView. Bioinformatics. 2010;26:3125–6.
Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–3.
Leo S, Gaïa N, Ruppé E, Emonet S, Girard M, Lazarevic V, Schrenzel J. Detection of bacterial pathogens from Broncho-alveolar lavage by next-generation sequencing. Int J Mol Sci. 2017;18(9):2011.
Lazarevic V, Gaïa N, Girard M, et al. When bacterial culture fails, metagenomics can help: a case of chronic hepatic Brucelloma assessed by next-generation sequencing. Front Microbiol. 2018;9:616.
Tossios P, Karatzopoulos A, Tsagakis K, et al. Treatment of infected thoracic aortic prosthetic grafts with the in situ preservation strategy: a review of its history, surgical technique, and results. Heart Lung Circ. 2014;23:24–31.
Wilson WR, Bower TC, Creager MA, et al. Vascular graft infections, mycotic aneurysms, and endovascular infections: a scientific statement from the American Heart Association. Circulation. 2016;134:e412–60.
Ducasse E, Calisti A, Speziale F, Rizzo L, Misuraca M, Fiorani P. Aortoiliac stent graft infection: current problems and management. Ann Vasc Surg. 2004;18:521–6.
Molacek J, Treska V, Baxa J, Certik B, Houdek K. Acute conditions caused by infectious Aortitis. Aorta (Stamford). 2014;2:93–9.
Heysell SK, Hughes MA. Listeria monocytogenes endovascular graft infection. Open Forum Infect Dis. 2016;3(1):ofv203–2.
Silvestri V, Isernia G. Suspected giant cell aortitis: from multiple aortic structural damage to fatal Listeria Sepsis, a case report. Ann Vasc Surg. 2017;42:307.e1–6.
Brossier J, Lesprit P, Marzelle J, Allaire E, Becquemin JP, Desgranges P. New bacteriological patterns in primary infected Aorto-iliac aneurysms: a single-Centre experience. Eur J Vasc Endovasc Surg. 2010;40:582–8.
Murphy K, Al-Jundi W, Nawaz S. Mycotic aneurysms of the abdominal aorta due to Listeria monocytogenes. Int J Surg Case Rep. 2013;4:626–8.
Infrarenal endoluminal bifurcated stent graft infected with Listeria monocytogenes. J Vasc Surg 1999;29:554–556.
Cernohorsky P, Reijnen MM, Tielliu IF, van Sterkenburg SM, van den Dungen JJ, Zeebregts CJ. The relevance of aortic endograft prosthetic infection. J Vasc Surg. 2011 Aug;54:327–33.
Rohde H, Horstkotte MA, Loeper S, et al. Recurrent Listeria monocytogenes aortic graft infection: confirmation of relapse by molecular subtyping. Diagn Microbiol Infect Dis. 2004;48:63–7.
Saleem BR, Berger P, Zeebregts CJ, Slart RH, Verhoeven EL, van den Dungen JJ. Periaortic endograft infection due to Listeria monocytogenes treated with graft preservation. J Vasc Surg. 2008;47:635–7.
Tanner-Steinmann B, Boggian K. Vascular Endograft infection with Listeria monocytogenes treated with surgical debridement but without graft removal. Case Rep Med. 2011;2011:482815.
Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a web browser. BMC Bioinformatics. 2011;12:385.
Acknowledgements
The authors are grateful to Dr. M Abbas for his critical reading of the manuscript.
Funding
None.
Availability of data and materials
The datasets analysed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
AF: work design, data acquisition, work draft; MC: work design, data acquisition, analysis and interpretation, work draft and revision; AC: data acquisition, analysis and interpretation; VL: data acquisition, analysis and interpretation, work revision; NG: data acquisition, analysis and interpretation; SL: data acquisition, analysis and interpretation, work revision; MG: data acquisition, analysis and interpretation, work revision; DM: data acquisition and analysis; JS: work design, data analysis and interpretation, work revision, approval of submitted version. All authors have approved the submitted version (and any substantially modified version that involves the author’s contribution to the study) and to have agreed both to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Written informed consent to publication was obtained from the patient, as well as written informed consent to store and analyse the aortic tissues and to use the results for research.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Foulex, A., Coen, M., Cherkaoui, A. et al. Listeria monocytogenes infectious periaortitis: a case report from the infectious disease standpoint. BMC Infect Dis 19, 326 (2019). https://doi.org/10.1186/s12879-019-3953-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-019-3953-z