Abstract
Background
Control of gonorrhea in resource-limited countries, such as Indonesia, is mostly unsuccessful. Examining Neisseria gonorrhoeae (Ng) transmission networks using strain typing might help prioritizing public health interventions.
Methods
In 2014, urogenital Ng strains were isolated from clients of sexually transmitted infection clinics in three Indonesian cities. Strains were typed using Multiple-Locus Variable Number Tandem Repeat (VNTR) Analysis (MLVA) and Ng Multi-Antigen Sequence Typing (NG-MAST) at the Public Health Service, Amsterdam, the Netherlands, and compared to Dutch strains collected from 2012 to 2015. Minimum spanning trees (MSTs) were constructed using MLVA profiles incorporating demographics and NG-MAST genogroups. A cluster was defined as ≥5 strains differing in ≤1 VNTR locus. The concordance between MLVA and NG-MAST was examined with the adjusted Wallace coefficients (AW).
Results
We collected a total of 78 Indonesian strains from Yogyakarta (n = 44), Jakarta (n = 25), and Denpasar (n = 9). Seven MLVA clusters and 16 non-clustered strains were identified. No cluster was specific for any geographic area, risk group or age group.
Combined with 119 contemporary Dutch strains, 8 MLVA clusters were identified, being four clusters of Indonesian strains, two of Dutch strains, and two of both Indonesian and Dutch strains. Indonesian strains (79.5%) were more often clustered compared to Dutch strains (24.3%).
The most prevalent NG-MAST genogroups among Indonesian strains was G1407 (51.3%) and among Dutch strains was G2992 (19.3%). In Indonesian strains, the AW [95% confidence interval] for MLVA to NG-MAST was 0.07 [0.00–0.27] and for NG-MAST to MLVA was 0.03 [0.00–0.12].
Conclusion
Indonesian Ng strains are more often clustered than Dutch strains, but show no relation with geographical area, risk group, or age group, suggesting a more clonal Ng epidemic in Indonesia. Some similar Ng strains circulate in both Indonesia and the Netherlands.
Similar content being viewed by others
Background
The prevalence of gonorrhea in Indonesia is high, especially among sex workers, men who have sex with men [MSM] and transwomen [1, 2]. For a successful control of gonorrhea and other sexually transmitted infections (STIs), especially in resource-limited countries, such as Indonesia, identification of high risk groups that need priority is required [3]. However, the overall risk to contract gonorrhea in the population is largely determined by the prevalence within transmission networks and the network characteristics [3,4,5].
In Indonesian STI clinics, transmission networks are traditionally identified by contact tracing using self-reported sexual history [6]. However, this approach is limited by recall bias, socially desirable responses, and privacy issues [7].
The use of molecular typing techniques may improve the quality and scope of transmission network identification by providing information on the spread of bacterial strains in different populations [8]. Various methods have been introduced to classify Neisseria gonorrhoeae (Ng) strains based on genotype [8,9,10].
Multiple-Locus Variable Number Tandem Repeat Analysis (MLVA) is a fast and robust method for molecular typing of Ng strains which is based on the variation in the number of tandem repeated DNA sequences in five loci of the bacterial genome [8, 9]. Ng Multi-Antigen Sequence Typing (NG-MAST) is a method using the combination of allele sequence variations of two gonococcal genes, porB and tbpB, to determine the strain type [8, 10].
No Indonesian Ng epidemiological study using MLVA or NG-MAST has yet been performed. We studied the transmission network of Ng in Indonesia by analyzing the distribution of Ng strains identified by MLVA and NG-MAST among clients of STI clinics in three major cities in Indonesia. In addition, we examined how Indonesian strains relate to other Ng strains by comparison with strains from Amsterdam, the Netherlands.
Methods
Study settings, population, and sample collection
This study was part of a larger study on gonorrhea epidemiology in Indonesia, which was partly reported elsewhere [2].
In 2014, clients of eight STI service facilities in three major cities in Indonesia (Jakarta, Yogyakarta, and Denpasar) were invited to participate in the study. A written informed consent was obtained from all participants. All participants were 16 years old or older at the day of recruitment. The participants completed a self-administered questionnaire regarding demographics, behavioral and clinical information. Based on the questionnaire data, three risk groups were defined: heterosexual males, heterosexual females, and a combined group of men who have sex with men and transwomen (MSM).
The clinicians collected urethral samples from males and transwomen, and endocervical samples from females. Primary culture and simple identification were performed at the laboratory of the Faculty of Biology, Universitas Gadjah Mada in Yogyakarta, Indonesia. Isolates were transferred on dry ice to the reference laboratory of the Public Health Service of Amsterdam, the Netherlands, where re-culture and complete identification were performed, as previously described [2]. The pure cultured Ng isolates were stored at −80 °C in beads (Microbank®, BioTrading, Mijdrecht, the Netherlands).
MLVA and NG-MAST have been used to type Ng strains at the reference laboratory in the past decade [5, 9]. A database containing molecular typing data of the strains, as well as characteristics of the patients from whom the strains were collected, was built in BioNumerics® version 7.5 (Applied-Maths, Austin, Texas, USA). To understand how Indonesian strains relate to circulating Ng strains in another setting, we used Dutch strains collected at the STI clinic of Amsterdam, the Netherlands, from two molecular epidemiology studies on azithromycin resistance [11], and ceftriaxone resistance [12]., All Dutch strains were fully susceptible for tested antibiotics as they acted as control strains in the two studies. The Dutch strains were collected between January 2012 and December 2015.
Molecular typing using MLVA and NG-MAST
DNA lysates were prepared by boiling one stored bead seeded with cultured Ng isolates in 100 μL of phosphate buffered saline at 95 °C for 15 min.
MLVA was performed as described [9]. In short, DNA lysates were used for two different multiplex PCRs in a Bio-Rad C1000 PCR System (Bio-Rad, Hercules, California, USA) to amplify five variable number of tandem repeat sequence (VNTR) loci, i.e. VNTR04–03, VNTR04–10, VNTR07–02, VNTR30–01 and VNTR16–01. The PCR products were diluted 1:20 in water and 2 μl of each diluted sample was mixed with 18 μl of a 1:450 in water diluted GeneScan LIZ 500 size standard (Applied Biosystems, Foster City, California, USA). After heat denaturation for 5 min at 95 °C, the fragments were separated on an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, California, USA) using the fragment analysis module. Sizing and calculation of the number of repeats of each VNTR were performed with GeneMarker® version 1.8 (SoftGenetics, LLC, State College, Pennsylvania, USA). An MLVA profile was a string consisting of integers that corresponds to the number of repeats found in the five VNTR loci [9]. This string was used for cluster analysis.
NG-MAST was also performed, as described [10]. In short, DNA lysates were used for two different PCR targets to amplify parts of the porB and tbpB genes in the Bio-Rad C1000 PCR System. The size of the amplimers in each PCR product was estimated using a QIAxcel® Advanced System (Qiagen, Hilden, Germany). PCR products with sufficient amplimers were transferred to the sequencing facility of the Academic Medical Center, University of Amsterdam, the Netherlands. An online international database of NG-MAST (www.ng-mast.net) was used as reference to determine allele numbers of porB and tbpB gene fragments, and to assign sequence types (STs). In addition, we checked the susceptibility profile of Indonesian strains against cephalosporins (ceftriaxone and cefixime), as previously reported [2], and examined its distribution by NG-MAST types.
Genogrouping, cluster analysis, and statistics
We built a BioNumerics® database integrating participants’ characteristics and molecular typing data of Indonesian and Dutch reference strains.
Minimum spanning trees (MSTs) were constructed using MLVA profiles [9]. Two MSTs were constructed, i.e. an MST using data of Indonesian strains only and another MST using combined data of both Indonesian and Dutch strains. A cluster was defined as a group of ≥5 strains that differed in maximally one VNTR locus [9]. Clusters were assigned capital letters.
Based on NG-MAST STs, a genogroup was defined among strains that had one identical allele (porB or tbpB), and had another allele differing in ≤4 base pairs (bp) (tbpB) or ≤5 bp (porB), as described [13]. Genogroups were named after the most frequent STs within that genogroup. A genogroup consisting of ≥5 strains was considered as major genogroup.
Various participants’ characteristics, i.e. geographical location (city of the recruiting clinic), risk group, and age group, and NG-MAST genogroups were indicated in the MSTs.
We also examined the concordance between MLVA and NG-MAST by calculating the adjusted Wallace coefficient (AW) and its 95% confidence interval (CI) [14], using an online tool (www.comparingpartitions.info). For each country of strains collection, the AWs were calculated if the number of strains that belonged to one of MLVA clusters and one of the major NG-MAST genogroups was large >(20 strains) [14]. The AW of MLVA to NG-MAST showed the probability of strains within an MLVA cluster to belong to the same major NG-MAST genogroup. The AW of NG-MAST to MLVA showed the probability of strains within the same NG-MAST genogroup to belong to the same MLVA cluster. The AWs range from 0.00 (no association between the two techniques) to 1.00 (a complete association).
The statistical analyses were performed in STATA version 13 (Stata Corp., College Station, Texas, USA).
Results
Characteristics and distribution of Indonesian ng strains
From the 992 participants recruited in Indonesia, a total of 78 strains were successfully typed at the reference laboratory. The strains were from Yogyakarta (n = 44, 56.4%), Jakarta (n = 25, 32.1%), and Denpasar (n = 9, 11.5%) (Table 1). Strains were from heterosexual males (n = 5, 6.4%), heterosexual females (n = 30, 38.5%), and MSM (n = 43, 55.1%). Most of the participants (n = 46, 59.0%) were sex workers. Strains were mostly from participants aged 25–34 years (n = 34, 43.6%).
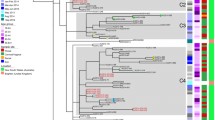
Regarding the Indonesian strains, seven clusters (cluster A-G) and 16 non-clustered strains were identified (Fig. 1 and Additional file 1: Table S1). Cluster A (n = 16) was the largest cluster. Cluster C was relatively close to cluster D, differing in only two integers of their MLVA profiles. Other clusters differed in three or more integers from each other (Fig. 1).
A minimum spanning tree (MST) based on Multiple-Locus Variable Number Tandem Repeat Analysis (MLVA) profile describing distribution of Neisseria gonorrhoeae (Ng) strains collected from Yogyakarta, Jakarta, and Denpasar, Indonesia in 2014. A circle represents strains that had identical MLVA profile; A gray halo surrounding circles represents a cluster, which was defined as a group of five or more strains that had a difference in at most one variable number tandem repeat (VNTR) locus, assigned capital letters (A-G); A solid line connecting circles represents a difference in one integer of MLVA profile; A dashed line connecting circles represents a difference in two integers of MLVA profile; *A grouping based on Ng Multi-antigen Sequence Typing (NG-MAST) (see Material and Methods section); a distribution of Indonesian Ng strains by city; b distribution of Indonesian Ng strains by risk group; c distribution of Indonesian Ng strains by age group; d distribution of Indonesian Ng strains by NG-MAST genogroup
Clusters B, C, E and G predominantly consisted of strains from Yogyakarta, whereas clusters A and F predominantly consisted of strains from Jakarta. In clusters A, B, and F, we observed strains from all three cities and no cluster was fully specific to one geographical area (Fig. 1a). Clusters A, B, and G predominantly consisted of strains from MSM and cluster F was predominantly from heterosexual females (Fig. 1b). In clusters C, D, and E, we observed strains from all risk groups, however (Fig. 1b). Clusters B and D predominantly consisted of strains from participants aged 25–34 years (which constituted the largest age group), but in clusters E and G, we observed strains from participants of all four age groups (Fig. 1c).
The 16 non-clustered strains were mostly isolated from Yogyakarta (n = 10, 62.5%) and from MSM (n = 11, 68.8%). The non-clustered strains were: three small groups each consisting of three strains with an identical MLVA profile, one group of two strains with an identical MLVA profile, and five singletons.
The most frequently occurring NG-MAST genogroup was G1407 (51.3%, Table 1) which was found in all Indonesian cities. We found no correlation between NG-MAST genogroups and geographical area, risk group, or age group (data not shown).
Relation between Indonesian and Dutch ng strains
We included 119 Dutch contemporary strains as reference strains (Table 1). Most were from MSM (n = 103, 86.6%), and the remainder were from heterosexual males (n = 6, 5.0%) and heterosexual females (n = 10, 8.4%). Strains from participants who aged 25–34 years (n = 39, 32.8%) composed the largest group, as was the case for the Indonesian participants.
Regarding both Indonesian and Dutch strains, we identified eight clusters, including the seven clusters that had been identified in the MST using Indonesian data only (Fig. 2, Table 2). Cluster C/D was a consolidation of cluster C and D found in MST using Indonesian data only, and one Dutch singleton. Two clusters (B and C/D) consisted of both Indonesian and Dutch strains. Two clusters (H and I) consisted of Dutch strains only. We also identified 118 non-clustered strains (16 Indonesian and 102 Dutch strains).
A minimum spanning tree (MST) based on Multiple-Locus Variable Number Tandem Repeat Analysis (MLVA) profile describing distribution of and relation between Neisseria gonorrhoeae (Ng) strains collected from Yogyakarta, Jakarta, and Denpasar, Indonesia (2014) and from Amsterdam, the Netherlands (2012–2015). A circle represents strains that had identical MLVA profile; a gray halo surrounding circles represents a cluster, which was defined as a group of five or more strains that had a difference in at most one variable number tandem repeat (VNTR) locus, assigned capital letters (a-i); a solid line connecting circles represents a difference in one integer of MLVA profile; a dashed line connecting circles represents a difference in two integers of MLVA profile
Indonesian strains (n = 64, 79.5%) were more likely to be included in a cluster than Dutch strains (n = 17, 14.3%, p < 0.001). An identical MLVA profile of all strains in a cluster was observed in six out of seven Indonesian clusters, whereas the two Dutch clusters showed VNTR differences within the cluster, suggesting more diversity in the Dutch clusters.
The three most frequent NG-MAST genogroups among Indonesian strains were G1407 (n = 40, 51.3%), G2992 (n = 13, 16.7%), and G9276 (n = 10, 12.8%). Four Indonesian strains with por allele 908 and tbpB allele 2413 had an unidentified ST, yet these strains could be included in genogroup G1407. Indonesian strains that belong to ST1407 did not show decreased susceptibility against cephalosporins (ceftriaxone and cefixime), as we previously reported [2]. NG-MAST data were available for 112 out of 119 Dutch strains (NG-MAST was unsuccessful in 7 strains). The three most frequent genogroups among Dutch strains were G2992 (n = 23, 19.3%), G21 (n = 15, 12.6%), and G5624 (n = 9, 7.6%) (Table 1).
In 55 Indonesian strains that could be included in an MLVA cluster and a major NG-MAST genogroup, the AW [95% CI] for MLVA to NG-MAST was 0.07 [0.00–0.27] and for NG-MAST to MLVA was 0.03 [0.00–0.12]. The AWs in Dutch strains were not calculated because of a small sample size (n = 13 strains).
Discussion
Here we describe Ng strains isolated from individuals living in three major Indonesian cities. A clustered pattern of Ng strains with no specific distribution regarding geographical location, risk group, or age group was observed. In addition, we found that most Indonesian strains were different from Dutch strains circulating in Amsterdam, the Netherlands, within overlapping time periods. Indonesian strains were more often clustered and therefore more clonal.
Several pathogen and individual factors may explain our findings. First, the finding of similar strain types circulating across different groups may be caused by the strain virulence and transmissibility [15, 16]. Strains with a higher level of virulence, e.g. those showing antimicrobial resistance or other high virulence characteristics, are more likely to survive and circulate more widely in the population [17,18,19,20,21]. Strains with resistance to third generation cephalosporins were so far not found in Indonesia [2], not even among ST1407 strains. This is in contrast to the situation in Europe, where fully susceptible ST1407 strains do occur, but the modal MIC of ST1407 strains to ceftriaxone is markedly increased [13].
Host and network factors also contribute to the pattern of strains circulating in the population [3, 4]. The finding of clusters that had no specific geographical location, risk group and age group distribution, may be associated with a disassortative sexual mixing which would enlarge the transmission network. Disassortative sexual mixing may be observed among individuals living in urban settings or among bridging populations (e.g. men who have sex with both men and women) [22,23,24,25].
A number of strains from patients in Indonesia were identified by active case finding in risk groups, whereas strains from Amsterdam were cultured from patients visiting the outpatient clinic. Active case finding may lead to identification of more epidemiologically related patients, and may in part explain the high frequency of clustering among Indonesian strains. However, this does not explain why clusters comprise strains from different cities.
Concordance between MLVA and NG-MAST was very low in Indonesian strains, much lower than was reported in previous studies [26, 27]. The number of studies comparing both MLVA and NG-MAST to type Ng strains is limited however, and the reported level of concordance between the two techniques varied greatly [26,27,28]. This could be caused by the different genetic regions of Ng targeted by these techniques [8]. Mutations or genetic translocation in these regions probably occur independently [15, 16]. In addition, discrepancy in the MLVA/NG-MAST concordance might relate to varying sample size and sample selection among studies [8, 26,27,28]. Also this MLVA has been validated using mainly European strains; it could be that by addressing different targets for Indonesian strains, this concordance could be improved.
MLVA is a relatively fast and cheap method compared to NG-MAST but needs a good local database. NG-MAST is a more robust technique with an internationally available database. To get maximum discriminatory typing power whole genome sequencing will be the best solution for typing but so far, pricing is still prohibitive certainly for developing countries [29].
We also observed a small overlap in MLVA clusters among Indonesian and Dutch strains. This could be the result of either an international spread of Ng strains or convergent evolutionary process that occurred in different geographical areas [15, 16].
Our study has some limitations. Indonesian and Dutch strains were collected through two distinct approaches. Data collection in Indonesia was performed only in 2014 (varying from 1 month to 5 months per participating clinic) and was focused on several specific risk groups, i.e. MSM, transwomen, and female sex workers living in an urban setting. Success of initial culture and of re-culture in the reference laboratory may be dependent on genetic background of the strains and on the timing of sample processing. For instance, strains from Yogyakarta, where the sample processing was quickly performed, were largely overrepresented (>56%). Therefore, our group of strains may not be totally representative for the Indonesian population under study.
We could not fairly compare the concordance of MLVA and NG-MAST between Indonesian and Dutch strains. The number of Dutch strains that could be included in one of the MLVA clusters and one of the major NG-MAST genogroups was too small for calculating the AWs. Small samples (n ≤ 20) results in wider 95% CI and lead to greater uncertainty in estimating the AWs [14].
The Dutch reference strains were all cultured immediately after sample collection from Ng positive visitors of only one STI clinic in Amsterdam, the Netherlands. The strains were collected regardless of risk group or age group criteria, yet the large majority of the strains (>87%) were nevertheless from MSM. These biases limit the comparison of Indonesian and Dutch strain data and limit the generalization of our findings to the population level.
Our study also has several strengths as it is the first to describe molecular epidemiology of Ng in Indonesia covering major cities and risk groups. It is also one of the few studies using both MLVA and NG-MAST techniques to describe Ng epidemiology. Finally, our study is able to show the relations of Ng strains circulating in two remotely separated settings (Indonesia and the Netherlands).
Conclusion
We describe clusters of Ng strains collected from individuals living in an Indonesian urban setting and compared them with strains circulating in Amsterdam, the Netherlands. Indonesian Ng strains were more often clustered than Dutch strains, but showed no specific relation with geographical area, risk group, or age group. This finding suggests that a limited number of Ng clonal complexes circulated throughout Indonesia, at least in the three major cities in this study. This observation may challenge the intention to prioritize individuals of a certain geographical area, risk group, or age group in the sexual health program, e.g. the antimicrobial susceptibility screening. To describe the actual situation in the population, proportional samples from each group might be required in that program.
By comparing Indonesian and Dutch data, most of the strains appear to be unique for each country. However, some overlap in the MLVA clusters suggests that identical Ng strains might circulate in both Indonesia and the Netherlands. Whether this is an international spread or caused by a convergent evolutionary process, cannot be determined here.
Finally, the finding of a low concordance between MLVA and NG-MAST could be an important consideration regarding selection of typing techniques for future studies. With a better availability and affordable pricing, the application of whole-genome sequencing to investigate Ng transmission and antimicrobial resistance can be considered as an option [29].
Abbreviations
- AW:
-
the adjusted Wallace coefficient
- CI:
-
confidence interval
- DNA:
-
deoxyribonucleic acid
- MLVA:
-
multi-locus variable number tandem repeat analysis
- MSM:
-
men who have sex with men
- MST:
-
minimum spanning tree
- Ng:
-
Neisseria gonorrhoeae
- NG-MAST:
-
Neisseria gonorrhoeae multi-antigen sequence typing
- PCR:
-
polymerase chain reaction
- ST:
-
sequence type
- STI:
-
sexually transmitted infections
- VNTR:
-
variable number tandem repeat
References
Ministry of Health Republic of Indonesia – Directorate General of Disease Control and Environmental Health. In: Integrated biological and behavioral survey 2013. Bahasa Indonesia: Jakarta: Kemenkes RI; 2013.
Hananta IP, van Dam AP, Bruisten SM, et al. Gonorrhea in Indonesia: high prevalence of asymptomatic urogenital gonorrhea but no circulating extended spectrum cephalosporins-resistant Neisseria gonorrhoeae strains in Jakarta, Yogyakarta, and Denpasar, Indonesia. Sex Transm Dis. 2016;43:608–16.
Steen R, Wi TE, Kamali A, et al. Control of sexually transmitted infections and prevention of HIV transmission: mending a fractured paradigm. Bull World Health Organ. 2009;87:858–65.
Risau-Gusman S. Influence of network dynamics on the spread of sexually transmitted diseases. J R Soc Interface. 2012;9:1363–72.
Heymans R, Matser A, Bruisten SM, et al. Distinct Neisseria gonorrhoeae transmission networks among men who have sex with men in Amsterdam, The Netherlands. J Infect Dis. 2012;206:596–605.
Ministry of Health Republic of Indonesia – Directorate General of Disease Control and Environmental Health. In: National guideline in the management of sexually transmitted infection. Bahasa Indonesia: Jakarta: Kemenkes RI; 2011.
Althaus CL, Turner KM, Mercer CH, et al. Effectiveness and cost-effectiveness of traditional and new partner notification technologies for curable sexually transmitted infections: observational study, systematic reviews and mathematical modelling. Health Technol Assess. 2014;18:1–100. vii-viii
Unemo M, et al. Review and recommendation or international methods for typing Neisseria gonorrhoeae isolates and their implications for improved knowledge or gonococcal epidemiology, treatment, and biology. Clin Microbiol Rev. 2011;24:447–58.
Heymans R, Schouls LM, van der Heide HGJ, et al. Multiple-locus variable-number tandem repeat analysis of Neisseria gonorrhoeae. J Clin Microbiol. 2011;49:354–63.
Martin IM, Ison CA, Aanensen DM, et al. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J Infect Dis. 2004;189:1497–505.
Wind CM, Bruisten SM, Schim van der Loeff MF, et al. A case-control study of molecular epidemiology in relation to azithromycin resistance in Neisseria gonorrhoeae isolates collected in Amsterdam, the Netherlands, between 2008 and 2015. Antimicrob Agents Chemother. 2017;61(6). pii: e02374–e02316.
Dam AP, Wind CM, Dierdorp M, et al. Reduced susceptibility to ceftriaxone in Neisseria gonorrhoeae in the Netherlands is now mainly found in association with an A501V/T mutation in the penA gene. Proceedings of the STI & HIV World Congress (Joint Meeting of the 22nd ISSTDR and the 18th of IUSTI); 2017 Jul 9–12; Rio de Janeiro, Brazil.
Chisholm SA, Unemo M, Quaye N, et al. Molecular epidemiological typing within the European Gonococcal Antimicrobial Resistance Surveillance Programme reveals predominance of a multidrug-resistant clone. Euro Surveill. 2013;18:pii: 20358.
Severiano A, Pinto FR, Ramirez M, et al. Adjusted Wallace coefficient as a measure of congruence between typing methods. J Clin Microbiol. 2011;49:3997–4000.
Maiden MC. Population genomics: diversity and virulence in the Neisseria. Curr Opin Microbiol. 2008;11:467–71.
Marri PR, Paniscus M, Weyand NJ, et al. Genome sequencing reveals widespread virulence gene exchange among human Neisseria species. PLoS One. 2010;5(7):e11835.
Kushnir AV, Ilina EN, Malakhova MV, et al. Multiple-locus variable number tandem repeat analysis of Neisseria gonorrhoeae isolates in Russia. Infect Genet Evol. 2013;14:8–14.
Heymans R, Bruisten SM, Golparian D, et al. Clonally related Neisseria gonorrhoeae isolates with decreased susceptibility to the extended-spectrum cephalosporin cefotaxime in Amsterdam, the Netherlands. Antimicrob Agents Chemother. 2012;56:1516–22.
Chen SC, Yin YP, Dai XQ, et al. First nationwide study regarding ceftriaxone resistance and molecular epidemiology of Neisseria gonorrhoeae in China. J Antimicrob Chemother. 2016;71:92–9.
Morita-Ishihara T, Unemo M, Furubayashi K, et al. Treatment failure with 2 g of azithromycin (extended-release formulation) in gonorrhoea in Japan caused by the international multidrug-resistant ST1407 strain of Neisseria gonorrhoeae. J Antimicrob Chemother. 2014;69:2086–90.
Nguyen D, Gose S, Castro L, et al. Neisseria gonorrhoeae strain with reduced susceptibilities to extended-spectrum cephalosporins. Emerg Infect Dis. 2014;20:1211–3.
Hertog S. Heterosexual behavior patterns and the spread of HIV/AIDS: the interacting effects of rate of partner change and sexual mixing. Sex Transm Dis. 2007;34:820–8.
van Veen MG, Kramer MA, Op de Coul EL, et al. Disassortative sexual mixing among migrant populations in The Netherlands: a potential for HIV/STI transmission? AIDS Care. 2009;21:683–91.
Gorbach PM, Murphy R, Weiss RE, et al. Bridging sexual boundaries: men who have sex with men and women in a street-based sample in Los Angeles. J Urban Health. 2009;86(Suppl 1):63–76.
Yun K, JJ X, Reilly KH, et al. Prevalence of bisexual behaviour among bridge population of men who have sex with men in China: a meta-analysis of observational studies. Sex Transm Infect. 2011;87:563–70.
Heymans R, Golparian D, Bruisten SM, et al. Evaluation of Neisseria gonorrhoeae multiple-locus variable-number tandem-repeat analysis, N. gonorrhoeae multiantigen sequence typing, and full-length porB gene sequence analysis for molecular epidemiological typing. J Clin Microbiol. 2012;50:180–3.
Kushnir AV, Muminov TA, Bayev AI, et al. Molecular characterization of Neisseria gonorrhoeae isolates in Almaty, Kazakhstan, by VNTR analysis, Opa-typing and NG-MAST. Infect Genet Evol. 2012;12:570–6.
Abu-Rajab K, Palmer HM, Scoular A, et al. To what extent does Neisseria gonorrhoeae multiantigen sequence typing of gonococcal isolates support information derived from patient interviews? Int J STD AIDS. 2009;20:414–7.
De Silva D, Peters J, Cole K, et al. Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study. Lancet Infect Dis. 2016;16:1295–303.
Acknowledgements
We would like to express our greatest appreciation to men, women, and transwomen participating in this study. We would also like to thank: the health care workers in the participating clinics, Non-Governmental Organizations (NGOs) staffs, and The Neisseria Project research assistants who contributed in data collection phase in Indonesia; laboratory technicians and management of the Public Health Service of Amsterdam; and the Indonesian government (the Ministry of Research, Technology and Higher Education and the Ministry of Health) for a good cooperation in supporting this research project.
Funding
This study was funded by the Beasiswa Unggulan Program, Ministry of Research, Technology, and Higher Education, Republic of Indonesia and the Public Health Service of Amsterdam, the Netherlands.
Availability of data and materials
The data that support the findings of this study are available from IPYH but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of IPYH.
Author information
Authors and Affiliations
Contributions
IPYH, AVD, MSVDL, HDV, and SMB contributed to the design of the study. IPYH coordinated data collection in Indonesia. IPYH, CMW and MD performed laboratory procedure for molecular typing and database building. IPYH performed statistical analyses. IPYH, AVD, MD, MSVDL, HDV, and SMB contributed in determining the direction of cluster analysis and performed interpretation of the results. IPYH prepared the manuscript draft. IPYH, AVD, CMW, MD, MSVDL, HDV, HS and SMB contributed in revising the manuscript critically for important intellectual content. SMB contributed in giving final approval of the version to be published. All authors agreed to be accountable for all aspects of the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The Medical and Health Research Ethics Committee (MHREC), Faculty of Medicine Universitas Gadjah Mada approved this study (Letter number: #KE/FK/38/EC). Written informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1: Table S1.
Characteristics and Distribution of Neisseria gonorrhoeae in Minimum Spanning Tree Constructed Based on Multiple-Locus Variable Number Tandem Repeat Analysis (MLVA) Profile of Strains Collected from Indonesian Major Cities in 2014 (DOCX 25 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hananta, I., van Dam, A.P., Schim van der Loeff, M.F. et al. Molecular epidemiology of Neisseria gonorrhoeae strains circulating in Indonesia using multi-locus variable number tandem repeat analysis (MLVA) and Neisseria gonorrhoeae multi-antigen sequence typing (NG-MAST) techniques. BMC Infect Dis 18, 7 (2018). https://doi.org/10.1186/s12879-017-2940-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-017-2940-5