Abstract
Background
In 2011–2012, Northern Vietnam experienced its first large scale hand foot and mouth disease (HFMD) epidemic. In 2011, a major HFMD epidemic was also reported in South Vietnam with fatal cases. This 2011–2012 outbreak was the first one to occur in North Vietnam providing grounds to study the etiology, origin and dynamic of the disease. We report here the analysis of the VP1 gene of strains isolated throughout North Vietnam during the 2011–2012 outbreak and before.
Methods
The VP1 gene of 106 EV-A71 isolates from North Vietnam and 2 from Central Vietnam were sequenced. Sequence alignments were analyzed at the nucleic acid and protein level. Gene polymorphism was also analyzed. A Factorial Correspondence Analysis was performed to correlate amino acid mutations with clinical parameters.
Results
The sequences were distributed into four phylogenetic clusters. Three clusters corresponded to the subgenogroup C4 and the last one corresponded to the subgenogroup C5. Each cluster displayed different polymorphism characteristics. Proteins were highly conserved but three sites bearing only Isoleucine (I) or Valine (V) were characterized. The isoleucine/valine variability matched the clusters. Spatiotemporal analysis of the I/V variants showed that all variants which emerged in 2011 and then in 2012 were not the same but were all present in the region prior to the 2011–2012 outbreak. Some correlation was found between certain I/V variants and ethnicity and severity.
Conclusions
The 2011–2012 outbreak was not caused by an exogenous strain coming from South Vietnam or elsewhere but by strains already present and circulating at low level in North Vietnam. However, what triggered the outbreak remains unclear. A selective pressure is applied on I/V variants which matches the genetic clusters. I/V variants were shown on other viruses to correlate with pathogenicity. This should be investigated in EV-A71. I/V variants are an easy and efficient way to survey and identify circulating EV-A71 strains.
Similar content being viewed by others
Background
Hand, foot and mouth disease (HFMD) is an acute febrile illness in children with a papulovesicular skin rash at the palms or soles of the feet, or both. Presentation can be with or without inclusion of mouth ulcers. Although the disease is usually mild and self-limiting, in some cases HFMD can result in severe complications such as encephalitis, aseptic meningitis, pulmonary edema, myocarditis, and death [29]. HFMD is caused by members of Human Enterovirus A, a family of picornaviridae which includes Coxsackievirus A (CV-A) and Human Enterovirus 71 (EV-A71) [3, 7]. The EV-A71 viruses are genetically related to CV-A; indeed, it has been suggested that these viruses may have diverged as recently as the 1940s [27]. Both EV-A71 and CV-A infections have been associated with severe HFMD in young children, sometimes resulting in death [2, 21, 33].
Enteroviruses are characterized by the presence of 4 structural proteins, VP4 being the internal capsid protein and VP1, VP2 and VP3 making the three external capsid proteins [6]. VP1 is the most external and is the main component of the canyon on the surface of picornaviruses. VP1 is involved in viral pathogenicity, receptor binding and immune modulation of EV-A71 [13, 31]. Differences in EV-A71 strains might contribute to the different severity of the disease [18, 24] and virulence determinants have been identified in the VP1 protein such as residues G/Q/R at position VP1–145, E at VP1–164 [5, 12, 16]. VP1 is used to classify enteroviruses. Based on the VP1 gene, EV-A71 is classified into three independent genogroups: A, B, and C. The EV-A71 B and C genogroups are each further subdivided into five subgenogoups, B1 to B5 and C1 to C5 [4].
Although EV-A71 was isolated for the first time in Vietnam in 2003, the first outbreak of HFMD was not reported in the southern provinces until 2005. The 2005 outbreak was associated with EV-A71 C1, C4 and C5 genotypes and Coxsackievirus A16 [14, 28]. In 2011, a major HFMD epidemic was reported in South Vietnam with fatal cases reported [19]. This 2011–2012 outbreak was the first one to occur in North Vietnam providing grounds to study the etiology, origin and dynamic of the disease. We report here the analysis of the VP1 gene of strains isolated throughout North Vietnam during the 2011–2012 outbreak.
Methods
Epidemiological information and source of specimens
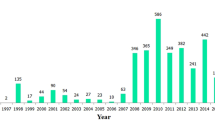
All HFMD cases in Northern provinces were reported to the National Institute of Hygiene and Epidemiology (NIHE) through the national communicable disease surveillance system since 2011. HFMD patients that reported to health centers or hospitals were diagnosed and classified in 4 severity levels (Additional file 1: Table S1). The evaluation of the disease was performed according to the guidelines specifically published by the Vietnamese Ministry of Health which are based on WHO and Taiwanese guidelines [11, 29]. A hundred and eight EV-A71 throat swabs from North Vietnam and 2 from Central Vietnam were collected from 2003 to 2012.
Sampling
Ninety four samples were obtained from 94 different hospitalized patients diagnosed with EV A71 HFMD in 19 out of 28 provinces in North Vietnam in 2011 and 2012 and stored at −80 °C. Fourteen reference samples obtained from previous cases of EV A71 HFMD between 2003 and 2010 in seven provinces in North Vietnam and two provinces in Central Vietnam were included in the study (Table 1). All samples were sent to the Enterovirus Laboratory of NIHE for etiological assays. Enterovirus-positive and EV-A71-positive samples were identified according to Nix et al. [20] using SO (SO224/SO222), AN (AN88/AN89) and MAS (MAS01S/MAS02A) [22] primer sets. Viral RNA was directly extracted from throat swab using QIAamp® Viral RNA Mini Kit (Qiagen, Valencia, USA). The cDNA was prepared using the GoScript™ Reverse Transcriptase kit from Promega. Seminested RT-PCR was conducted as described by Nix et al. [20]. The cDNA was first synthesized from the RNA for 10 min at 25 °C and followed by synthesis of the second strand at 42 °C for 50 min, 72 °C for 15 min using primers AN32, AN33, AN34 and AN35 [20]. PCR was done as described by Nix et al. [20] with 40 cycles of amplification (95 °C for 30 s, 42 °C for 30 s and, 60 °C for 45 s). One microliter of the first PCR was used a second seminested amplification for 40 amplification cycles of 95 °C for 30 s, 60 °C for 20 s, and 72 °C for 15 s. Sequencing was performed with the Sanger method using the is Bigdye Terminator V3.1 cycle sequencing kit from Applied Biosystems in an ABI sequencer 3130.
Sequence analyses
Sequences were deposited in GenBank and accession numbers are provided in Table 1. The VP1 genetic sequences were aligned in Seaview 4.6 [10] using Muscle algorithm [9]. Best-fitting evolutionary models were determined by JModelTest 2.1 [8] or by ProtTest 2.4 [1] using the corrected version of the Akaike Information Criterion (AICc). The phylogeny of VP1 was performed by Maximum Likelihood (ML) inference using the model GTR + G + F with Seaview 4.6 [10]. The robustness of nodes was assessed with 500 bootstrap replicates. ML analysis of the amino acid sequences was performed using the model JTT + I + G with Seaview 4.6 [10]. The robustness of nodes was assessed with 500 bootstrap replicates. Sequence polymorphism was investigated using the DnaSP 5.10.01 package [17]. Amino acid numbering was done with respect to full length polyprotein using the sequence HQ129932.1 [16] as a reference.
Bias and ethics
Training session on HFMD cases definition and reporting were organized for the staff of the routine surveillance system to enhance quality and consistency of case report. This work was conducted following the requirements of the Vietnamese Ministry of Health and under the Law of Communicable Diseases Prevention and Control passed in 2007.
Factorial correspondence analysis
A factorial correspondence analysis (FCA) was performed using XLSTAT software (Addinsoft®). Variables considered were: amino acid profiles on positions 151, 164 and 186 (V/I), respectively in this work (249, 262 and 284 on the full length VP1 protein), severity level, ethnicity, age of patients, and patient location. The best descriptive axes were retained, explaining 34% of the data spread. Potential correlations were confirmed by a test of contingency using Statview 5.0 software (SAS Institute Inc.)
Spatio-temporal analysis
Administration data were obtained from GADM database of Global Administrative Areas (version 2.8, November 2015). Spatial analyses were conducted with Quantum GIS, version 2.8.2. All spatial data are in the WGS 84 coordinate system.
Results
Clinical and epidemiological features
Data are shown in Table 1. Patients age ranged from 2 months to 12 years old (median at 1.8 years, IQR of 1.5 years). 102/106 (96.23%) patients were under 5. The age-specific incidence highest in the 1–2 years age group (44 cases, 41.51%) and remained very low for older children. The lowest incidence was observed in infants under 6 months (2.83%) and children above 10 years old (0.94%). Patients came from all parts of the region including mountainous, rural and urban areas. Out of 83 cases, 59 (71.08%) belonged to main Vietnamese ethnicity (Ethnicity 1) while the rest of patients belonged to the minority Hmong ethnicity (Ethnicity 2) (Table 1). All severity levels were reported for the patients. Mild forms (severity level 1) made the majority of cases (57 cases, 61.29%) while 15 patients displayed severe symptoms (16.13%). Among this group, 3 patients displayed a severity score of 3. No case with the highest level of 4 was recorded. Moderate forms of HFMD were found in 21 patients (22.58). (Table 1).
VP1 phylogeny and population structure
Phylogenetic analysis of the VP1 gene sequences indicated the presence of four clusters (Fig. 1). Cluster 1, the closest to the root, was the main one, comprising 56 sequences and structured into several subclusters characterized by low bootstrap values. Cluster 2 and cluster 3 comprised respectively 23 and 12 sequences and segregated from cluster 1 to which they were separated with a low bootstrap of 30. The last cluster, cluster 4, comprising 17 sequences was characterized by a high bootstrap value (100) and was a derivate from cluster 3. A similar tree topology was observed when using protein alignments (Data not shown). With respect to the current genogroup classification, clusters 1, 2 and 3 belonged to the subgenogroup C4 whereas cluster 4 belonged to the subgenogroup C5 (Table 1). The VP1 protein was characterized by a high rate of conservation. However, three sites displayed a consistent Valine (V) / Isoleucine (I) variation (Fig. 2). These sites, sites A, B and C were located in the VP1 protein at position 814, 827 and 849 of the polyprotein. Six types of I/V variants were observed when considering the amino acids at sites A, B and C: VVI (1 sequence), IVI (15 sequences), IIV (28 sequences), VII (38 sequences), VIV (23 sequences) and VVV (3 sequences) (Table 1). When attributing a color code for each I/V variant population and applying it to the phylogenetic tree a clear overlap with the previously detected clusters was observed (Fig. 1). Cluster 4 overlapped with the IVI population while clusters 2 and 3 comprised the VII variants. The VVV, IVV and VIV variants all corresponded to cluster 1. However they did not mix and each one corresponded to a specific subcluster. The VVI variant derived from the cluster 4 / IVI group. Three exceptions were found, with VII variants present in clusters 1 and 4. With respect to phylogeny, the I/V variant closest to the root is VVV, the VIV population emerged from this group and gave rise in turn to the IIV group. The VII groups, derived from the VIV group with first cluster 2 from which cluster 3 evolved. The well separated cluster 4/IVI variants evolved from cluster 3. This overlap between clusters and I/V populations was even stronger at the protein level since almost all the variability was borne by the I/V mutations, the rest of the protein being highly conserved. Groups VII, IIV, VVV and VIV belonged to the subgenogroup C4 whereas groups IVI and VVI belonged to the subgenogroup C5.
Nucleic acid polymorphism
When considering the polymorphism of the various clusters identified, very different traits were observed (Table 2). Cluster 1 was polymorphic (θ = 14.58) but with a 2.5 times more parsimony informative sites than singletons and 10-times more synonymous mutations than non-synonymous ones, suggesting that it is not a recent polymorphism or expanding population. The Ka/Ks ratio was also characterized by a low value. Conversely, cluster 2 displayed a very low level of polymorphism with a q of 4.06 and a low number of mutations η (η = 15). The Ka/Ks was slightly higher at 0.107. This is suggesting the existence of a bottleneck at the origin of cluster 2. Cluster 3 which originated from cluster 2 was more polymorphic with a slightly increasing θ (θ = 6.92) and number of mutations η (η = 19). Only synonymous mutations were observed resulting in turn in a very low Ka/Ks ratio of 0.029. Cluster 4 was on the other hand displaying a very high polymorphism with a η of 113 for 109 and a very high level of synonymous mutations (105 out of 110) suggesting a strong negative selection acting on a mutating population. As a consequence, the Ka/Ks ratio was also very low at 0.011.
Distribution of mutations and correlation analysis
The correlation analysis indicated a partial structuration of cases (34% of dispersion explained) on different parameters (Fig. 3). The VVI variant was not included in the analysis because all information was not available. The analysis was therefore conducted on only five variants, i.e. IIV, VII, IVI, VIV and VVV. Severity of HFMD seemed to correlate with the age of patient (p = 0.011) and the highest severity level was not observed above 11-month old. The VII variant segregated from the other variants on the F1 axis and was associated with both low severity (p = 0.025) and with the ethnicity-2 group, 56.5% of patients from this ethnicity-2 group were infected by the VII variant, but this represented only 46.4% of all samples harboring the VII protein (p = 0.006). No variant was specifically associated with the highest severity (p = 0.99) whereas the IIV variant was correlated with mild severity (p = 0.003).
Spatiotemporal distribution of the virus populations
I/V variants present in the 2011 outbreak belonged mostly the IIV and VII populations which were already present in Northern Vietnam (Fig. 4a) The IIV population was previously detected in 2008 in the Ninh Binh province whereas VII variants were detected in Cao Bang and Hai Phong in 2007 and in Nam Dinh and Ninh Binh 2008. VII and IIV variants represented 46% and 33.3% of the samples collected in 2011, respectively (Fig. 4b, d). Other mutant populations detected in 2011 were: IVI (1.7%) already detected in 2007 in Yen Bai and Han Nam, in 2008 in Ninh Binh and in 2010 in Hai Phong and Bac Kan; VIV (14.3%) previously found in 2003 in Ha Noi and in 2006 in Phu Yen; and VVV (4.8%) (Fig. 4a). The VVV variants were not detected in samples collected prior to the 2011–212 outbreak. The mutant populations detected in 2012 were IIV and VII whose prevalence was reduced to 16.2% and 19.3% and VIV and IVI which prevalence rose to 38.7% and 25.8%, respectively (Fig. 4c, d). The VVV mutant was found only in 2011 in Thanh Hoa, Nam Dinh and Ha Noi (Fig. 4b, d). With respect to spatial distribution, the rise of variants VII and IIV observed in 2011 was not located in a specific area but covered most of the sampling sites (8 out of 11). The replacement of the IIV and VII variants by the IVI and VIV variants followed a similar pattern confirming the wide-spread diffusion of the outbreak. The number of sites with more than two variants was higher in 2011 than in 2012. The IIV variant was the most widely spread in 2011 but became the least widely spread in 2012 (Fig. 4b, c). Conversely, the IVI variant which was the least widely spread in 2011 and found in only one province, i.e. Hoa Binh, became the most widely spread in 2012. The VVV variant was found only in 2011 in three provinces, in the South Eastern part of North Vietnam each time in association with the IIV variant (Fig. 4b, c). The VVI variant was detected only in Quang Nam, Central Vietnam and prior to 2011.
Discussion
This work provides an insight on the evolution and dynamics of the EV-A71 enterovirus during the first outbreak recorded in North Vietnam in 2011–2012. The first conclusion is that the 2011–2012 outbreak in North Vietnam was not due to a single exogenous strain imported from South Vietnam where HFMD outbreaks were present [19] or from another region. All variant populations observed during the 2011–2012 outbreak were already present in North Vietnam. The only exception is the VVV population which was found only in 2011 in three different provinces. However, the phylogenetic analysis indicated that this VVV variant was the closest to the root and therefore to the mother and oldest population. The reason for the lack of VVV variants in samples older than 2011 is most likely related to the low number of samples and to the low prevalence of this population. Furthermore, this 2011–2012 outbreak was also characterized by the cocirculation of the same four variant populations with a replacement between 2011 and 2012. The VII and IIV variants which were the most prevalent in 2011 were replaced by IVI and VIV populations in 2012. There is no clear explanation for the replacement of the main variant populations between 2011 and 2012 but it could be related to immunoresistance acquired during the first half of the outbreak in 2011 The surge of variants VII and IIV in the first part of the epidemic could not be related to any measured parameters and altogether the question remains of what triggered the outbreak in 2011 although all virus populations were already present. All I/V populations present at the beginning of the outbreak were capable of triggering it as shown by the replacement in 2012. It is not related either to the subgenogroup since the populations which emerged in 2012 belonged to two different subgenogroups, the VIV variant belonging the subgenogroup C4 and the IVI variant being a member of subgenogroup C5. A partial explanation could be a differential susceptibility of the human population which could have been slightly more susceptible to the VII and IIV groups. Another explanation might be found in the spatial distribution of the various variant groups, the socio-economic pattern and the route of dissemination. This work was not structured to address this issue and specific sampling schemes as well as transversal analyses should thus be further undertaken.
Another main outcome of this work is the observed correlation between I/V variant groups and phylogeny, pathogenicity and ethnicity. One hypothesis is that fixation of mutations in VP1 could be related to the VP1 function itself. Li et al. [16] reported virulent determinants in VP1 located at position 710 and 729 with Glutamic acid, Glycine or Arginine being associated to severe cases at position 710 and Glutamic acid at position 729 being also a marker of severity. In this study we didn’t see this correlation with position 710 bearing 90 Glutamic acid, 6 Glycine and 12 Glutamine and position 729 bearing 107 Aspartic acid and 1 Glycine both out of 108 sequences. These amino acids were found in strains associated to mild and moderately severe cases. No highly severe case was found in this work. I/V groups, although based on the relative arrangement of only three amino-acids, overlap the different clusters identified. These clusters correspond to genetically different populations characterized by specific polymorphism traits. This overlap between the specific combination of I/V residues at three positions and the phylogeny established on the nucleotide sequences suggests the occurrence of a selective pressure on the I/V arrangement. The high conservation of the proteins, despite variability at the nucleotide level, indicative of a negative, or purifying, selection pressure, indicates that the clustering at the protein level is driven by the I/V arrangement. The remaining question is what is the selective pressure acting on I/V variants and what could be the role of these I/V mutations. I/V mutations are located in the VP1 protein at positions 814, 827 and 849 of the polyprotein. The region from amino acid 697 to 862 on the EV-A71 polyprotein at the level of the VP1 protein was shown to be crucial for increasing the strength of protein-protein interactions in the capsid and its stability. This increased stability strongly enhances the pathogenicity and survival of the virus in the gastrointestinal track [15]. Isoleucine and valine are aliphatic hydrophobic amino acid mediating the core structure of the protein and have been reported to be involved in virulence and pathogenicity in several viruses, coxsackievirus B3 [25], Moloney murne Leukemia Virus [26], Infectious Bursal Disease virus [32], Japanese Encephalitis virus [30], and Simian-Human Immunodeficiency Virus [23]. The recurrent reports of the involvement of isoleucine and valine in the viral pathogenicity process in different viruses as well as their involvement in the selective pressure applied on the EV-A71 isolates analyzed in this work suggest that the I/V pattern at positions 249, 262 and 284 on the VP1 protein might indeed play a role in pathogenicity. The observed correlation of I/V variant populations with severity and ethnicity strengthen this hypothesis. However, the ethnicity correlation could be a result of spatial structuration since ethnicity-2 is mostly present in the Hòa Bình province. This in turn would suggest that the various EV-A71 variants display a geographic specificity.
Conclusion
Altogether, these data suggest that EV-A71 strains could remain in a low level, asymptomatic state, in genomic stasis and with a geographic structuration. The cause for outbreaks should thus be sought for in the socio-economic patterns rather than in exogenous emergence. Further investigations are needed to investigate this hypothesis and to bring valuable information for the management of this major pediatric disease.
References
Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–5.
Abu Bakar S, Chee HY, Al-Kobaisi MF, Xiaoshan J, Bing CK, Kit LS. Identification of enterovirus 71 isolates from an outbreak of hand, foot and mouth disease (HFMD) with fatal cases of encephalomyelitis in Malaysia. Virus Res. 1999;61:1–9.
Ang LW, Koh BK, Chan KP, Chua LT, James L, Goh KT. Epidemiology and control of hand, foot and mouth disease in Singapore. Ann Acad Med Singap. 2009;38:106–12.
Brown BA, Oberste MS, Alexander JP Jr, Kennett ML, Pallansch MA. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J Virol. 1999;73:9969–75.
Caine EA, Moncla LH, Ronderos MD, Friedrich TC, Osorio JE. A single mutation in the VP1 of Enterovirus 71 is responsible for increased virulence and Neurotropism in adult interferon-deficient mice. J Virol. 2016;90:8592–604.
Carter J, Saunders VA. Virology: principles and applications. Hoboken: John Wiley & Sons; 2007.
Chen KT, Chang HL, Wang ST, Cheng YT, Yang JY. Epidemiologic features of hand-foot-mouth disease and herpangina caused by enterovirus 71 in Taiwan, 1998–2005. Pediatrics. 2007;120:e244–52.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772.
Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics. 2004;5:113.
Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4.
Huang CC, Liu CC, Chang YC, Chen CY, Wang ST, Yeh TF. Neurologic complications in children with Enterovirus 71 infection. N Engl J Med. 1999;341:936–42.
Huang SW, Tai CH, Fonville JM, Lin CH., Wang SM, Liu CC, .Su IJ, Smith DJ, Wang JR. Mapping enterovirus A71 antigenic determinants from viral evolution. J Virol 2005; 89:11500-11506.
Kataoka C, Suzuki T, Kotani O, Iwata-Yoshikawa O, Nagata N, Ami Y, Wakita T, Nishimura Y, Shimizu H. The role of VP1 amino acid residue 145 of Enterovirus 71 in viral fitness and pathogenesis in a Cynomolgus monkey model. PLoS Pathog. 2015;11(7):e1005033.
Khanh TH, Sabanathan S, Thanh TT, Thoa le PK, Thuong TC, Hang VT, Farrar J, Hien TT, Chau NV, van Doorn HR. Enterovirus 71-associated hand, foot, and mouth disease, southern Vietnam, 2011. Emerging Infect Dis. 2012;18:2002–5.
Lal SK, Kumar P, Yeo WM, Kar-Roy A, Chow VT. The VP1 protein of human enterovirus 71 self-associates via an interaction domain spanning amino acids 66–297. J Med Virol. 2006;78:582–90.
Li R, Zou Q, Chen L, Zhang H, Wang Y. Molecular analysis of virulent determinants of enterovirus 71. PLoS One. 2011;6(10):e26237.
Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2.
McMinn P, Lindsay K, Perera D, Chan HM, Chan KP, Cardosa MJ. Phylogenetic analysis of enterovirus 71 strains isolated during linked epidemics in Malaysia, Singapore, and Western Australia. J Virol. 2001;75:7732–8.
Nguyen NTB, Pham H, Hoang CQ, Nguyen TM, Nguyen LT, Phan HC, Phan LT, Vu LN, Minh NNT. Epidemiological and clinical characteristics of children who died from hand, foot and mouth disease in Vietnam, 2011. BMC Infect Dis. 2014;14:341.
Nix WA, Oberste MS, Pallansch MA. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol. 2006;44:2698–704.
Ooi MH, Wong SC, Lewthwaite P, Cardosa MJ, Solomon T. Clinical features, diagnosis, and management of enterovirus 71. The Lancet Neurology. 2010;9:1097–105.
Perera D, Podin Y, Akin W, Tan CS, Cardosa MJ. Incorrect identification of recent Asian strains of Coxsackievirus A16 as human enterovirus 71: improved primers for the specific detection of human enterovirus 71 by RT PCR. BMC Infect Dis. 2004;4:11.
Peyerl FW, Barouch DH, Yeh WW, Bazick HS, Kunstman J, Kunstman KJ, Letvin NL. Simian-human immunodeficiency virus escape from cytotoxic T-lymphocyte recognition at a structurally constrained epitope. J Virol. 2003;77:12572–8.
Sanders S, Herrero L, McPhie K, Chow S, Craig M, Dwyer D, Rawlinson W, McMinn PC. Molecular epidemiology of enterovirus 71 over two decades in an Australian urban community. Arch Virol. 2006;151:1003–13.
Schmidtke M, Hammerschmidt E, Schüler S, Zell R, Birch-Hirschfeld E, Makarov VA, Wutzler P. Susceptibility of coxsackievirus B3 laboratory strains and clinical isolates to the capsid function inhibitor pleconaril: antiviral studies with virus chimeras demonstrate the crucial role of amino acid 1092 in treatment. J Antimicrob Chemother. 2005;56:648–56.
Szurek PF, Yuen PH, Ball JK, Wong PK. A Val-25-to-Ile substitution in the envelope precursor polyprotein, gPr80env, is responsible for the temperature sensitivity, inefficient processing of gPr80env, and neurovirulence of ts1, a mutant of Moloney murine leukemia virus TB. J Virol. 1990;64:467–75.
Tee KK, Lam TTY, Chan YF, Bible JM, Kamarulzaman A, Tong C, Takebe Y, Pybus OG. Evolutionary genetics of human enterovirus 71: origin, population dynamics, natural selection, and seasonal periodicity of the VP1 gene. J Virol. 2010;84:3339–50.
Tu PV, Thao NTT, Perera D, Huu TK, Tien NTK, Thuong TC, How OM, Cardosa MJ, McMinn PC. Epidemiologic and virologic investigation of hand, foot, and mouth disease, southern Vietnam, 2005. Emerging Infect Dis. 2007;13:1733–41.
WHO. A Guide to Clinical Management and Public Health Response for Hand, Foot and Mouth Disease (HFMD). WHO WPRO; 2011.
Yamaguchi Y, Nukui Y, Tajima S, Nerome R, Kato F, Watanabe H, Kurane I. An amino acid substitution (V3I) in the Japanese encephalitis virus NS4A protein increases its virulence in mice, but not its growth rate in vitro. J Gen Virol. 2011;92:1601–6.
Yang SL, Chou YT, Wu CN, Ho MS. Annexin II binds to capsid protein VP1 of enterovirus 71 and enhances viral infectivity. J Virol. 2011;85:11809–20.
Yu F, Ren X, Wang Y, Qi X, Song J, Gao Y, Wang X. A single amino acid V4I substitution in VP1 attenuates virulence of very virulent infectious bursal disease virus (vvIBDV) in SPF chickens and increases replication in CEF cells. Virology. 2013;440:204–9.
Zeng M, Li YF, Wang XH, Lu GP, Shen HG, Yu H, Zhu QR. Epidemiology of hand, foot, and mouth disease in children in shanghai 2007–2010. Epidemiol Infect. 2012;140:1122–30.
Acknowledgements
Not applicable.
Funding
The work was supported by internal grants from NIHE and from DUKE/NUS for sequencing. NDN was in part supported by European Erasmus Mundus project MAHEVA and by the PEPS project MoDyCa from University of Montpellier and CNRS.
Availability of data and materials
All data are publicly available and sequences have been deposited in Genbank. Accession numbers of sequences deposited in Genbank are ranging From KX906261 to KX906368 (108 sequences).
Data described in this article are publicly and fully available. All data are described in tables within the manuscript and sequences have been deposited in Genbank. Accession numbers are provided in Table 1.
Authors’ contributions
NDN participated to all parts of the work. OMS, YAH, RC and DJG generated all sequences. LTTH, LTSH, VDT and NTHT participated to sample collection and molecular analysis and amplification. AA designed all maps and spatiotemporal analysis. LG, CM, PR, GK, EC and RF participated to all bioinformatic, statistic and phylogenetic analyses. CD, NTH and TND have provided fruitful advises and discussions. RF supervised the work and participated to all analyses and to the writing. All authors read and approved the final manuscript.
Competing interests
The authors declare that there is no competing interests.
Consent for publication
Not Applicable.
Ethics approval and consent to participate
This work was conducted strictly following the requirements of the Vietnamese Ministry of Health and under the Law of Communicable Diseases Prevention and Control passed in 2007. This work was conducted under the control of NIHE Ethic committee. These procedures include a written agreement from parents or their legal representatives.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Additional file
Additional file 1: Table S1.
Severity levels of HFMD cases according to guidelines from the Vietnamese Ministry of Health. (DOCX 15 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Duy, N.N., Huong, L.T.T., Ravel, P. et al. Valine/isoleucine variants drive selective pressure in the VP1 sequence of EV-A71 enteroviruses. BMC Infect Dis 17, 333 (2017). https://doi.org/10.1186/s12879-017-2427-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-017-2427-4