Abstract
Background
High-risk human papillomavirus (HR HPV) testing is already part of cervical cancer screening programs in a number of countries. New tests need to be validated not only in clinical studies but also in routine screening settings with regard to their clinical performance.
Methods
The Abbott RealTime High Risk HPV Test (RT hrHPV test) was evaluated in a random sample of 1,456 patients from a German routine screening population of 13,372 women ≥30 years of age screened primarily by liquid-based cytology (LBC) that was complemented by 48 CIN3+ cases. Clinical sensitivities, relative specificities and positive predictive values (PPV) for both HPV tests were determined based on histologically confirmed high-grade cervical disease (CIN3+) as clinical outcome.
Results
HR HPV prevalence in residual LBC samples was found to be 5.4 % by the RT hrHPV test and 5.6 % by the HR HC2 test, respectively. The Kappa-value for overall agreement between the RT hrHPV test and the HC2 assay for detection of HR HPV was 0.87. Relative sensitivities for detection of CIN3+ in patients with abnormal cytology was 93.8 % for the RT hrHPV assay and 97.9 % for HC2 (p-value = 0.5). Relative specificities and PPVs were comparable for both tests. The highest PPV was calculated for the specific detection of HPV16 by the RT hrHPV test (84.2 %). The RT hrHPV test showed a reduced sensitivity for detection of HVP31-positive CIN3 + .
Conclusion
The RT hrHPV assay is as sensitive and specific in detecting severe cervical lesions in women with abnormal cytology as the HC2 HR HPV test.
Similar content being viewed by others
Background
Since the introduction of opportunistic cytological screening in Germany in 1971 the cervical cancer mortality rate has notably decreased [1]. However, 4,600 new cases and approximately 1,500 deaths due to cervical cancer are diagnosed in Germany each year [2]. Moreover, 150,000 cases of cervical cancer precursors (CIN3) are detected annually [3] and cervical cancer is the cause for 1.5 % of all female cancer deaths in Germany [2]. As persistent infection with High-Risk Human Papillomaviruses (HR HPV) is a necessary risk factor for the development of (pre)-cancer, numerous HPV tests are nowadays commercially available [4] to be used in cervical cancer screening programs [5]. However, only a small number of these tests have been approved by the FDA [6] with the Digene Hybrid Capture 2 High-Risk HPV DNA test as the first one (HC2; QIAGEN Hilden, Germany). The HC2 has been developed for the collective detection of 13 carcinogenic HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68) [7]. It is one of the best validated HPV tests whose methodology is based on nucleic acid hybridization with signal amplification for qualitative detection of HPV-DNA within cervical samples.
The Abbott RealTime High Risk HPV Test (RT hrHPV) is another fully automated HPV DNA test which is based on real-time PCR that targets the L1 region of the 13 carcinogenic HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68 and for detection of HPV 66. The multiplex design of the assay allows HPV16 and 18 genotyping as well as the collective detection of the 12 remaining HPV types [8]. The RT hrHPV test has been fully validated by several cross-sectional studies that evaluated its clinical performance in referral populations [9–18]. However, to date only four screening population-based cross sectional studies [19–22] and one follow-up study are available [23]. The objective of this retrospective study was the cross-sectional evaluation of the RT hrHPV assay in a routine cervical cancer screening population comprising women aged ≥ 30 years in Germany.
Methods
The study cohort and methodology has previously been published [24].
Study design
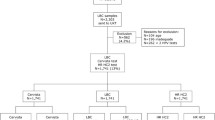
Briefly the study was conducted on a sample of 2,303 women of a routine screening population of 13,372 women ≥ 30 years of age living in the Hannover area of Germany in 2011 (Fig. 1). Cervical samples were collected in PreservCyt® Pap Test specimen collection medium (Hologic) and cytology was tested within 1 week after collection in a central services laboratory (Amedes, Bad Münder, Germany). After exclusion of ineligible samples, a total of 1,456 residual liquid based cytology (LBC) smears from this cohort including all samples with abnormal and ASC-US cytology results as well as 10 % of randomly selected samples with normal cytology results were tested by both HR HC2 and Abbott RealTime High Risk HPV Test (RT hrHPV). This collection was complemented with samples from 48 patients with CIN3+ from a separate referral cohort in order to obtain a sufficient CIN3+ rate. HC2 tests were conducted in the laboratory of the Division of Experimental Virology at the University Hospital in Tübingen. RT hrHPV assays were performed in the Amedes laboratory within 2 weeks after collection. Samples with discordant HPV test results were genotyped by INNO-LiPA HPV Genotyping Extra® for discrepancy analysis.
Samples were anonymized thus, participants were unaware of their HPV test results and colposcopy as well as histopathology was only performed when indicated by German standard operating procedures. Samples with a primary histology of ≥ CIN2+ were independently reviewed by up to two pathologists.
Sample collection
Sample Collection has previously been described [24]. Briefly cervical samples were collected in LBC PreservCyt® Collection medium (Hologic) using the Cervex broom according to routine guidelines. Samples were then centrally tested by cytology within one week of collection.
Liquid based cytology
LBC was performed as described before [24] by the ThinPrep® 2000 Processor (Hologic) following the manufacturer’s instructions. Cytology results were reported using the Munich Nomenclature II and were translated into The Bethesda System (TBS) ([25]; Table 1). LBC results were considered negative when the result was Pap I/II (Normal) or Pap IIw (ASC-US); all other results were considered positive [24].
HPV testing and genotyping
LBC samples were tested by Abbott RealTime High Risk HPV Test (RT hrHPV) in compliance with the manufacturer’s instructions. All specimens included in this study were initially tested by RT hrHPV test and subsequently analyzed by Digene Hybrid Capture 2 High-Risk HPV DNA (HC2) test. HC2 testing has previously been described [24].
HPV genotyping was carried out using the INNO-LiPA HPV Genotyping Extra® test (Fujirebio LiPA Extra) which identifies 20 HPV genotypes classified as Group 1, 2A and 2B carcinogens (16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 69, 70, 73, 82) and 8 low-risk HPV or intermediate risk genotypes (6, 11, 40, 43, 44, 54, 71, 74). LiPA Extra is a line blot assay based on SPF-10-PCR as described previously [26]. Strips were scanned and analyzed automatically with a flatbed scanner and the LiRAS software (Fujirebio).
Histology reviews
As detailed before all samples with an initial histology result of ≥ CIN2+ were reviewed by an independent external expert. In the case of a discrepant review reading, a second histology review was performed. If two out of three diagnoses were identical, the result was considered final [24].
Statistical analysis
As described [24] statistical analysis was performed on all samples with valid LBC, RT hrHPV and HC2 (N = 1,456) test results. To calculate the agreement between RT hrHPV and HC2, the Cohen’s kappa value (κ) was used. The Wilson score method was used to calculate 95 % confidence intervals (CI) for HPV prevalence. Moreover, clinical sensitivity and relative specificity as well as positive predictive values (PPV) and negative predictive values (NPV) were calculated according to Cuzick et al. [20] based CIN3+ histology results.
Relative performance of the two tests was measured by calculating the ratio of the sensitivity to the specificity. This ratio of the sensitivity of the two tests was defined as the True Positive rate of the first test divided by the True Positive rate of the second test. The relative specificity on the other hand, is dependent of prevalence, and was expressed as Spec(RT hrHPV)/Spec(HC2) = (0.818-Prevalence)/(0.800 - Prevalence). The delta method was used to determine confidence intervals. A full description of this method has previously been published [24]. Statistical analysis of the relative sensitivity and specificity was calculated using the statistics software package R version 3.0.2.
Results
Of 2,303 overall specimens a total of 847 samples had to be excluded from the analysis either because they returned unsatisfactory cytology results (n = 196), because they could not be analyzed by all three tests due to insufficient material (n = 547) or because study participants were younger than 30 years of age (n = 104). The 1,456 remaining specimens were tested by both RT hrHPV and HC2 (Fig. 1). 358 of these samples had an abnormal cytology result (Pap ≥ III; ≥ ASC-H) 56 were classified as ASC-US (Pap IIw) and 1,042 women had normal cytology results (Pap I/II) (Table 1 and 2). Colposcopy and histopathology on biopsies was only performed on 33 patients with abnormal cytology results independent of the HPV test results. A total of 9 CIN2 cases were identified by review histopathology. The remaining 24 biopsies were < CIN2. In order to obtain a higher CIN3+ rate, a total of 48 CIN3+ cases were added to the cohort.
HPV prevalence and type distribution
Concordant HPV DNA test results were obtained from 1,391 of 1,456 LBC samples (Table 2). Thus the overall percentage of agreement was 95.5 % and the Kappa coefficient was κ = 0.87 (95%CI: 0.84-0.90). The overall HR HPV positivity rate determined by HC2 was high with 23.4 % and 21.4 % by RT hrHPV. In the cytology normal group (Pap I/II) HR HPV was detected in 5.4 % of the specimens by RT hrHPV and in 5.6 % using the HC2 assay, respectively (Table 1 and Fig. 2). In the ASC-US category, HPV prevalence was 21.4 % for RT hrHPV and 28.6 % for HC2. In samples with abnormal cytology HR HPV was detected in 50 % of the LBC specimens classified as ASC-H, AGC (Pap III) by RT hrHPV as well as HC2, compared to 65.1 % versus 72.1 %, respectively, in the LSIL, HSIL (Pap IIID) category and 94.7 % (RT hrHPV) versus 100 % (HC2) in category HSIL, CIS (Pap IVa). HPV prevalence in women with glandular abnormal cytology (Pap III+; AGC+) was 68 % detected with RT hrHPV and 71.3 % with HC2 test.
Cohen’s Kappa Coefficients (κ) were calculated to measure the agreement of RT hrHPV and HC2 within the different LBC categories (Table 1). Kappa values (κ) were excellent for normal (Pap I/II) samples (κ = 0.78) and all abnormal LBC categories ≥ LSIL (≥Pap III κ = 0.78). For ASC-US (Pap IIw) specimens the agreement was fair (κ = 0.62).
A total of 65 discordant samples were detected between the RT hrHPV and HC2 (Table 3). 47 discrepant samples (72.3 %) were RT hrHPV-negative and HC2-positive compared to only 18 samples (27.7 %) with HC2-negative and RT hrHPV-positive test results. The majority of discordant results was detected in specimens with ASC-H (PapIII 14.3 %) cytology results followed by the LSIL, HSIL (Pap IIID, 10.3 %) and the normal (Pap I/II, 2.3 %) cytology categories. All deviant samples (n = 65) were genotyped using the INNO-LiPA HPV Genotyping Extra test in order to identify false positive or false negative samples (Table 3). As a result we resolved 5 RT hrHPV-negative and 7 HC2-negative samples as true negatives. While four samples were inadequate for LiPA genotyping, all remaining samples (n = 49) were HPV DNA positive.
In detail one sample missed by HC2 contained a non-target type of the HC2 assay and a total of 9 specimens (50 %) were false-negative by HC2. However, none of the samples missed by HC2 had a histology result of CIN3+. Interestingly, the HC2 assay detected HPV DNA in 18 RT hrHPV-negative samples, which contained non-target types of either HPV DNA test including 11 cases of HPV 53.
In contrast 18 discordant samples (38.3 %) with negative RT hrHPV and positive HC2 test results were non-target types of the RT hrHPV test (including 2 HPVX types), while another 21 samples (44.7 %) were false-negative by RT hrHPV revealing an unusually high false-negative rate for the RT hrHPV test. These 21 samples also included two specimens with CIN3+ histology. Both samples were found to be positive for high risk HPV type 31 by LiPA Extra genotyping with a total of six CIN3+ samples positive for this HPV type.
The RT hrHPV test concurrently detects 14 different HPV types and generates genotyping information for specific identification of the highly carcinogenic types HPV 16 and 18. The association of HPV 16 and HPV 18 with different cytology outcome as detected by RT hrHPV is shown in Fig. 2. The overall detection rate for HPV 16 (6.8 %) was higher than that of HPV 18 (1.0 %). Prevalence of HPV 16 and 18 increases proportionally with higher grade of cytology abnormalities and was highest in HSIL and CIS (≥Pap IVa). 58.5 % of these samples were positive for HPV 16 and 12.2 % for HPV 18. In contrast HPV 16 prevalence was 2.2 % compared to 0.2 % for HPV18 in the normal (Pap I/II) and 3.6 % versus 0 %, respectively, in the ASC-US (Pap IIw) category. HPV 16 and HPV 18 were both detected in 6.3 % of the specimens with ASC-H, AGC (Pap III) cytology results in contrast to the LSIL, HSIL (Pap IIID) category, which included 19.9 % HPV 16 positive and 2.3 % HPV 18 positive samples. All samples with a histology result of CIN3+ were genotyped by using the INNO-LiPA HPV Genotyping Extra test. Overall HR HPV genotype distribution in specimens with CIN3+ lesions is shown in Fig. 3. All HPV types detected in both single and multiple infections were included in the calculations. Not surprisingly, HPV 16 (46.2 %) was the most prevalent genotype in specimens with CIN3+ lesions followed by HPV 31 (12.3 %), HPV 52 (9.2 %), HPV 33 (6.2 %) and HPV 18 (4.6 %). All specimens with a LiPA Extra genotyping result of either HPV 16 or HPV18 were correctly identified by the concurrent Abbott RT HPV 16/18 genotyping.
Sensitivity and specificity
Relative sensitivity and specificity were calculated to define the diagnostic accuracy of the HR HC2 and the RT hrHPV test for detection of CIN3+. Within this cohort 81 biopsies were available from patients with abnormal cytology results. Overall relative sensitivity for detection of CIN3+ in patients with abnormal cytology was 93.8 % for the Abbott RealTime hrHPV assay and 97.9 % for HC2, a difference which is insignificant (p-value = 0.5; Table 4). Similar results were obtained for the endpoint CIN2+ (94.7 % vs 98.2 % respectively). Relative specificities and positive predictive values (PPV) were comparable 81.1 % and 16.9 % for RT hrHPV and 79.8 % and 16.5 % for HC2, respectively. The PPV for the detection of CIN2+ was highest at 84.2 % for the separate detection of HPV 16 by RT hrHPV. Overall 50 histology reviews were performed. Sensitivity and specificity results for the performances of both HPV tests with endpoint CIN3+ did not change when the review results were accounted for in the respective calculations (data not shown).
Colposcopy was given only to women with abnormal cytology findings; therefore the true number of high grade CIN in the normal (Pap I/II) and ASC-US (Pap IIw) categories is not known. Due to this lack of definitive data one could not calculate the clinical sensitivity and specificity of the two tests. However, we calculated the ratio of the sensitivity and specificity of the two tests to each other, thus we are able to determine relative, if not absolute, performance. The ratio of sensitivities between RT hrHPV test and HC2 is 0.959 (95%CI: 0.885–1.033). Fig. 4 shows the ratio of specificity as a function of prevalence. We found no statistical difference between clinical sensitivity and specificity of the RT hrHPV test and HC2 for the detection of high grade disease at the 95 % confidence level.
Discussion
This study was conducted to evaluate the analytical and relative performance of the RT hrHPV test using residual LBC specimens selected from a German routine cervical cancer screening population. HR HPV positivity rates overall (23.4 % for HC2 and 21.4 % for RT hrHPV) and in LBC normal samples (5.4 % for RT hrHPV and 5.6 % for HC2) were in line with previously published data [21]. HPV prevalence in women with abnormal cytology (AGC+ ≥ PapIII) was 68 % detected by RT and 71.3 % for the HC2 test. The generally slightly higher detection rates of the HC2 test may be attributed to cross-reactivity with non-target types [27]. The most prevalent HPV type in women with high grade cervical disease in this study was HPV 16, which is in accordance with all recent studies and meta-analyses (summarized by [28]).
Comparing the performances of both tests we showed that the RT hrHPV and HC2 test performed similarly and the agreement of both assays was excellent (κ = 0.87). These results reflect previously reported data [13, 19, 21] and indicate that both HPV DNA tests performed equivalently. Discordant samples were analyzed by the LiPA HPV genotyping Extra test, which has been used as an adjudicating assay in test comparison studies before [24, 29, 30], due to its high analytical sensitivity [31]. We found that 50 % of HC2-negative, but RT hrHPV-positive samples were true HPV negative. The possibility of false-negative HC2 results has previously been reported to be attributed to a low viral copy number leading to false-negative HC2 results [32] or to the lack of an internal control for cellularity, which is however ruled out here by the split sample protocol. Furthermore, we found that the HC2 test was able to detect infections with the non-target type HPV 53. Cross-reactivity of the HC2 test with HPV 53 has been demonstrated by multiple previous reports. In fact, HPV 53 has been shown to be one of the most frequent non-target types detected by the HC2 test through cross-hybridization of its HR probe [21, 33–35]. While the HC2 test detected all cases of CIN3+, the RT hrHPV test missed two CIN3+ cases positive for HPV 31. Similar results have previously been published by Poljak et al., who reported that the RT hrHPV test missed two CIN3 + −cases with HPV 31 and 58 co-infections [36]. Two other reports also demonstrated a diminished sensitivity of the RT hrHPV test for HPV 31-positive CIN3 cases [21, 30]. Indeed in the present study we found that one of the two CIN3+ cases missed by RT hrHPV test represented a co-infection with HPV-type 33, which might indicate competitive primer binding in the PCR leading to an unfavorable kinetic of amplification for HPV 31 or other types from the alpha-9 subgenus in mixed infections.
Comparing concurrent genotyping results of the RT hrHPV test we were able to show that RT HPV 16/18 genotyping correctly identified all specimens with a LiPA Extra genotyping result of either HPV 16 or HPV 18. These results are in contrast with earlier reports showing concordance between LiPA and RT genotyping in only 90 % of the tested specimens [10].
Relative sensitivities for detection of CIN3+ were high and comparable for both tests (p-value = 0.5) whereas relative specificities for RT was slightly higher than for the HR HC2 test. By calculating the ratios of sensitivities and specificities, respectively, we were able to confirm that no statistical difference exists between the two tests’ performances. Our observations are in line with cross-sectional studies reported previously in routine screening populations [19–21] and suggest that RT hrHPV test is well suited to be used in routine primary cervical cancer screening or adjunctive to cytology. Further evidence for the applicability of the RT hrHPV test in primary screening has recently been published by Poljak et al., reporting the first longitudinal data for the Abbott RT HPV test [23]. The authors demonstrated the non-inferior clinical performance of the RT HPV test in a routine screening population of 3,920 women with a 3-year follow-up time in comparison to the performance of the HC2 test.
In summary we found that the sensitivities and relative specificities of the RT hrHPV and the HR HC2 test are comparable. However, it appears that the RT hrHPV test has a reduced sensitivity for the detection of HPV type 31, which in this study has led to two CIN3+ cases missed by the RT hrHPV test. Because HPV 31 is one of the most prevalent high-risk HPV-types worldwide [37] and is found in 3,5 % of global cervical cancer cases [38], it is important to carefully assess risks and benefits of applying the RT hrHPV test. A considerable benefit of the RT hrHPV HPV test is its separate type specific detection of the most prevalent HPV genotypes 16 and 18, which together account for a total of an estimated 70.4 % of cervical cancer cases worldwide [38]; and we demonstrated that the PPV for detecting high grade disease is by far highest for Abbott RT hrHPV HPV 16.
Conclusions
We provided evidence that the Abbott HR HPV test is suitable for primary routine screening in Germany and other countries with a similar infrastructure regarding the secondary prevention of cervical cancer.
Abbreviations
- AGC:
-
Atypical glandular cells
- ASC-H:
-
Atypical squamous cells – cannot exclude HSIL
- ASC-US:
-
Atypical squamous cells of undetermined significance
- CI:
-
Confidence interval
- CIN:
-
Cervical intraepithelial neoplasia
- CIS:
-
Carcinoma in situ
- HC2:
-
Digene Hybrid Capture 2 High-Risk HPV DNA Test
- HPV:
-
Human Papillomavirus
- HR:
-
High-risk
- HSIL:
-
High grade squamous intraepithelial lesion
- LBC:
-
Liquid-based cytology
- LSIL:
-
Low grade squamous intraepithelial lesion
- NPV:
-
Negative predictive value
- PPV:
-
positive predictive value
- RLU/CO:
-
Relative light units/cutoff
- RT hrHPV test:
-
Abbott RealTime High Risk HPV Test
References
Schenck U, von Karsa L. Cervical cancer screening in Germany. Eur J Cancer. 2000;36(17):2221–6.
RKI: Krebs in Deutschland 2009/2010. Robert Koch-Institut und die Gesellschaft der epidemiologischen Krebsregister in Deutschland eV, Berlin 2013, 9. Ausgabe.
Siebert U, Sroczynski G, Hillemanns P, Engel J, Stabenow R, Stegmaier C, Voigt K, Gibis B, Holzel D, Goldie SJ. The German cervical cancer screening model: development and validation of a decision-analytic model for cervical cancer screening in Germany. Eur J Public Health. 2006;16(2):185–92.
Cuzick J, Bergeron C, von Knebel Doeberitz M, Gravitt P, Jeronimo J, Lorincz AT, C JLMM, Sankaranarayanan R, P JFS, Szarewski A: New technologies and procedures for cervical cancer screening. Vaccine. 2012;30(Suppl 5):F107–116
Poljak M, Kocjan BJ, Ostrbenk A, Seme K. Commercially available molecular tests for human papillomaviruses (HPV): 2015 update. J Clin Virol. 2016;76(Suppl 1):S3–S13.
Arbyn M, Snijders PJ, Meijer CJ, Berkhof J, Cuschieri K, Kocjan BJ, Poljak M. Which high-risk HPV assays fulfil criteria for use in primary cervical cancer screening? Clin Microbiol Infect. 2015;21(9):817–26.
Iftner T, Villa LL. Chapter 12: Human papillomavirus technologies. J Natl Cancer Inst Monogr. 2003;31:80–8.
Huang S, Tang N, Mak WB, Erickson B, Salituro J, Li Y, Krumpe E, Schneider G, Yu H, Robinson J, et al. Principles and analytical performance of Abbott RealTime High Risk HPV test. J Clin Virol. 2009;45 Suppl 1:S13–17.
Halfon P, Benmoura D, Agostini A, Khiri H, Penaranda G, Martineau A, Blanc B. Evaluation of the clinical performance of the Abbott RealTime high-risk HPV for carcinogenic HPV detection. J Clin Virol. 2010;48(4):246–50.
Venturoli S, Leo E, Nocera M, Barbieri D, Cricca M, Costa S, Santini D, Zerbini M. Comparison of Abbott RealTime High RiskHPV and hybrid capture 2 for the detection of high-risk HPV DNA in a referral population setting. J Clin Virol. 2012;53(2):121–4.
Jentschke M, Soergel P, Lange V, Kocjan B, Doerk T, Luyten A, Petry KU, Poljak M, Hillemanns P. Evaluation of a new multiplex real-time polymerase chain reaction assay for the detection of human papillomavirus infections in a referral population. Int J Gynecol Cancer. 2012;22(6):1050–6.
Huang S, Erickson B, Tang N, Mak WB, Salituro J, Robinson J, Abravaya K. Clinical performance of Abbott RealTime High RiskHPV test for detection of high-grade cervical intraepithelial neoplasia in women with abnormal cytology. J Clin Virol. 2009;45 Suppl 1:S19–23.
Cuzick J, Ambroisine L, Cadman L, Austin J, Ho L, Terry G, Liddle S, Dina R, McCarthy J, Buckley H, et al. Performance of the Abbott RealTime high-risk HPV test in women with abnormal cervical cytology smears. J Med Virol. 2010;82(7):1186–91.
Wong OG, Lo CK, Szeto E, Cheung AN. Efficacy of Abbott RealTime High RiskHPV test in evaluation of atypical squamous cells of undetermined significance from an Asian screening population. J Clin Virol. 2011;51(2):136–8.
Szarewski A, Mesher D, Cadman L, Austin J, Ashdown-Barr L, Ho L, Terry G, Liddle S, Young M, Stoler M, et al. Comparison of seven tests for high-grade cervical intraepithelial neoplasia in women with abnormal smears: the predictors 2 study. J Clin Microbiol. 2012;50(6):1867–73.
Veijalainen O, Tuomisaari S, Luukkaala T, Maenpaa J. High risk HPV testing in the triage of repeat ASC-US and LSIL. Acta Obstet Gynecol Scand. 2015;94(9):931–6.
Salimovic-Besic I, Tomic-Cica A, Smailji A, Hukic M. Comparison of the detection of HPV-16, 18, 31, 33, and 45 by type-specific DNA- and E6/E7 mRNA-based assays of HPV DNA positive women with abnormal Pap smears. J Virol Methods. 2013;194(1–2):222–8.
Halfon P, Benmoura D, Agostini A, Khiri H, Penaranda G, Martineau A, Blanc B. Stepwise algorithm combining HPV high-risk DNA-based assays and RNA-based assay for high grade CIN in women with abnormal smears referred to colposcopy. Cancer Biomarkers. 2010;7(3):133–9.
Carozzi FM, Burroni E, Bisanzi S, Puliti D, Confortini M, Giorgi Rossi P, Sani C, Scalisi A, Chini F. Comparison of clinical performance of Abbott RealTime High RiskHPV test with that of hybrid capture 2 assay in a screening setting. J Clin Microbiol. 2011;49(4):1446–51.
Cuzick J, Cadman L, Mesher D, Austin J, Ashdown-Barr L, Ho L, Terry G, Liddle S, Wright C, Lyons D, et al. Comparing the performance of six human papillomavirus tests in a screening population. Br J Cancer. 2013;108(4):908–13.
Poljak M, Ostrbenk A, Seme K, Ucakar V, Hillemanns P, Bokal EV, Jancar N, Klavs I. Comparison of clinical and analytical performance of the Abbott realtime High RiskHPV test to the performance of hybrid capture 2 in population-based cervical cancer screening. J Clin Microbiol. 2011;49(5):1721–9.
Hesselink AT, Meijer CJ, Poljak M, Berkhof J, van Kemenade FJ, van der Salm ML, Bogaarts M, Snijders PJ, Heideman DA. Clinical validation of the Abbott RealTime High RiskHPV assay according to the guidelines for human papillomavirus DNA test requirements for cervical screening. J Clin Microbiol. 2013;51(7):2409–10.
Poljak M, Ostrbenk A, Seme K, Sterbenc A, Jancar N, Vrtacnik Bokal E. Three-year longitudinal data on the clinical performance of the Abbott RealTime High Risk HPV test in a cervical cancer screening setting. J Clin Virol. 2016;769Suppl 1):S29–39.
Boehmer G, Wang L, Iftner A, Holz B, Haedicke J, von Wasielewski R, Martus P, Iftner T. A population-based observational study comparing cervista and hybrid capture 2 methods: improved relative specificity of the cervista assay by increasing its cut-off. BMC Infect Dis. 2014;14(1):674.
Petry KU, Menton S, Menton M, van Loenen-Frosch F, de Carvalho GH, Holz B, Schopp B, Garbrecht-Buettner S, Davies P, Boehmer G, et al. Inclusion of HPV testing in routine cervical cancer screening for women above 29 years in Germany: results for 8466 patients. Br J Cancer. 2003;88(10):1570–7.
Petry KU, Luyten A, Justus A, Iftner A, Strehlke S, Schulze-Rath R, Iftner T. Prevalence of low-risk HPV types and genital warts in women born 1988/89 or 1983/84 -results of WOLVES, a population-based epidemiological study in Wolfsburg. Ger BMC Infect Dis. 2012;12:367.
de Cremoux P, Coste J, Sastre-Garau X, Thioux M, Bouillac C, Labbe S, Cartier I, Ziol M, Dosda A, Le Gales C, et al. Efficiency of the hybrid capture 2 HPV DNA test in cervical cancer screening. A study by the french society of clinical cytology. Am J Clin Pathol. 2003;120(4):492–9.
Bruni L, Barrionuevo-Rosas L, Albero G, Aldea M, Serrano B, Valencia S, Brotons M, Mena M, Cosano R, Muñoz J et al.: Human Papillomavirus and Related Diseases in the World. ICO Information Centre on HPV and Cancer (HPV Information Centre) 2015, Summary Report.
Ngou J, Magooa MP, Gilham C, Djigma F, Didelot MN, Kelly H, Yonli A, Sawadogo B, Lewis DA, Delany-Moretlwe S, et al. Comparison of careHPV and hybrid capture 2 assays for detection of high-risk human papillomavirus DNA in cervical samples from HIV-1-infected African women. J Clin Microbiol. 2013;51(12):4240–2.
Hwang Y, Lee M. Comparison of the AdvanSure human papillomavirus screening real-time PCR, the Abbott RealTime High Riskhuman papillomavirus test, and the hybrid capture human papillomavirus DNA test for the detection of human papillomavirus. Ann Lab Med. 2012;32(3):201–5.
Torres M, Fraile L, Echevarria J, Hernandez Novoa B, Ortiz M. Human papillomavirus (HPV) genotyping: automation and application in routine laboratory testing. Open Virol J. 2012;6:144–50.
Jastania R, Geddie WR, Chapman W, Boerner S. Characteristics of apparently false-negative digene hybrid capture 2 high-risk HPV DNA testing. Am J Clin Pathol. 2006;125(2):223–8.
Gillio-Tos A, De Marco L, Carozzi FM, Del Mistro A, Girlando S, Burroni E, Frayle-Salamanca H, Giorgi Rossi P, Pierotti P, Ronco G, et al. Clinical impact of the analytical specificity of the hybrid capture 2 test: data from the New Technologies for Cervical Cancer (NTCC) study. J Clin Microbiol. 2013;51(9):2901–7.
Castle PE, Solomon D, Wheeler CM, Gravitt PE, Wacholder S, Schiffman M. Human papillomavirus genotype specificity of hybrid capture 2. J Clin Microbiol. 2008;46(8):2595–604.
Lindemann ML, Dominguez MJ, de Antonio JC, Sandri MT, Tricca A, Sideri M, Khiri H, Ravet S, Boyle S, Aldrich C, et al. Analytical comparison of the cobas HPV test with hybrid capture 2 for the detection of high-risk HPV genotypes. J Mol Diagn. 2012;14(1):65–70.
Poljak M, Kovanda A, Kocjan BJ, Seme K, Jancar N, Vrtacnik-Bokal E. The Abbott RealTime High RiskHPV test: comparative evaluation of analytical specificity and clinical sensitivity for cervical carcinoma and CIN 3 lesions with the hybrid capture 2 HPV DNA test. Acta Dermatovenerol Alp Panonica Adriat. 2009;18(3):94–103.
Bruni L, Diaz M, Castellsague X, Ferrer E, Bosch FX, de Sanjose S. Cervical human papillomavirus prevalence in 5 continents: meta-analysis of 1 million women with normal cytological findings. J Infect Dis. 2010;202(12):1789–99.
Clifford G, Franceschi S, Diaz M, Munoz N, Villa LL. Chapter 3: HPV type-distribution in women with and without cervical neoplastic diseases. Vaccine. 2006;24 Suppl 3:S3/26–34.
Acknowledgements
Publication of this manuscript was supported by the DFG-Open Access Publishing Fund of the University of Tübingen.
Funding
Not applicable.
Availability of data and materials
Anonymous data are available from the corresponding author on reasonable request.
Authors’ contributions
TI designed the study, was involved in data analysis and interpretation and drafted the manuscript. LW and PM performed the Ratio of specificities and sensitivities analysis and were involved in drafting the manuscript. BH was the studies coordinator, performed statistical analyses and drafted the manuscript. AI and NI performed HPV testing and genotyping. JHJ wrote the manuscript. RvW performed histologies. GB recruited study participants, collected the samples and performed LBC. All authors read and approved the final manuscript.
Competing interests
This study was supported by an unconditional grant from Hologic Incorporated to the University Hospital of Tübingen, which hosts the Division of Experimental Virology. Hologic had no influence on the study protocol, no access to primary data and was not involved in the analysis and writing of the manuscript.
Consent for publication
Not applicable.
Ethics approval and consent to participate
As determined by the ethics committee (Ethikkommission bei der Deutschen Ärztekammer Niedersachsen), no ethical approval or consent was required for this study, because the data was collected anonymously.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Iftner, T., Wang, L., Iftner, A. et al. Study-based evaluation of the Abbott RealTime High Risk HPV test in comparison to the HC2 HR HPV test in women aged ≥30 years using residual LBC ThinPrep specimens. BMC Infect Dis 16, 672 (2016). https://doi.org/10.1186/s12879-016-1994-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-016-1994-0