Abstract
Background
Rett syndrome (RTT) is a neurological disorder mainly caused by mutations in MeCP2 gene. It has been shown that MeCP2 impairments can lead to cytokine dysregulation due to MeCP2 regulatory role in T-helper and T-reg mediated responses, thus contributing to the pro-inflammatory status associated with RTT. Furthermore, RTT subjects suffer from an intestinal dysbiosis characterized by an abnormal expansion of the Candida population, a known factor responsible for the hyper-activation of pro-inflammatory immune responses. Therefore, we asked whether the intestinal fungal population of RTT subjects might contribute the sub-inflammatory status triggered by MeCP2 deficiency.
Methods
We evaluated the cultivable gut mycobiota from a cohort of 50 RTT patients and 29 healthy controls characterizing the faecal fungal isolates for their virulence-related traits, antifungal resistance and immune reactivity in order to elucidate the role of fungi in RTT’s intestinal dysbiosis and gastrointestinal physiology.
Results
Candida parapsilosis, the most abundant yeast species in RTT subjects, showed distinct genotypic profiles if compared to healthy controls’ isolates as measured by hierarchical clustering analysis from RAPD genotyping. Their phenotypical analysis revealed that RTT’s isolates produced more biofilm and were significantly more resistant to azole antifungals compared to the isolates from the healthy controls. In addition, the high levels of IL-1β and IL-10 produced by peripheral blood mononuclear cells and the mixed Th1/Th17 cells population induced by RTT C. parapsilosis isolates suggest the capacity of these intestinal fungi to persist within the host, being potentially involved in chronic, pro-inflammatory responses.
Conclusions
Here we demonstrated that intestinal C. parapsilosis isolates from RTT subjects hold phenotypic traits that might favour the previously observed low-grade intestinal inflammatory status associated with RTT. Therefore, the presence of putative virulent, pro-inflammatory C. parapsilosis strains in RTT could represent an additional factor in RTT’s gastrointestinal pathophysiology, whose mechanisms are not yet clearly understood.
Similar content being viewed by others
Background
The gut mycobiota, together with the bacterial microbiota, exerts key roles in maintaining the intestinal microbial community structure, metabolic functions and has strong immunomodulatory properties, being a main actor in host physiopathology [1]. The host response to fungi is mediated at first by the innate immunity through recognition of the fungal pathogen-associated molecular patterns (PAMPs) by the host cells’ pattern recognition receptors (PRRs). The C-type lectin receptors (CLRs; e.g. dectin 1 and dectin 2, also known as CLEC7A and CLEC6A respectively) are fundamental for fungal recognition and for the development of innate and adaptive immune responses, especially T-helper (Th) 1 and Th17 responses [2]. Th1 cells, through the production of IFNγ and TNFα, activates and recruits phagocytes (macrophages, neutrophils) at potential sites of infection [2], while Th17 cells are necessary for protection against fungal infections [3, 4]. In fact, IL-17 ability to mobilize neutrophils and to induce the production of antimicrobial peptides contributes to an efficient control of fungi at different body sites [2]. However, well balanced pro-inflammatory and tolerogenic responses are a prerequisite to avoid potential harmful inflammatory responses triggered by gastrointestinal fungi. The shift between pro-inflammatory and tolerogenic dendritic cells (DCs) responses are mediated by the kynurenine pathway of tryptophan catabolism, in which the expression of indoleamine 2,3-dioxygenase (IDO1) has a key role on plasticity of DCs activities in balancing between CD4+ effector Th cells and regulatory T (T-reg) cells [2, 5]. IDO1 is widely recognized as a regulator of immune homeostasis and suppressor of inflammation by inducing IL-10 through the production of immune-active kynurenines that activate the aryl-hydrocarbon receptor (AHR) in lymphoid tissues [6], thus inducing the transcription of FOXP3 and promoting immune tolerance via T-reg cells [7, 8]. Nevertheless, the mechanisms by which commensal fungi choose to shift their phenotype towards infection are not well understood, even if the disruption of the microbial community structure resulting in intestinal dysbiosis has been proposed to be one of the reasons [9]. Indeed, alterations of the gut microbiota can lead to inflammation involving hyper activation of Th1 and Th17 immune responses [10]. Altered immunological response to fungi can in turn contribute to systemic inflammatory responses. Remarkably, fungal infections shift IDO1’s activity [11, 12], reducing the levels of kynurenine and thus promoting inflammation [13]. Furthermore, alterations of the levels of kynurenine, a neuroprotective agent, have been implicated in several pathologies, including autism spectrum disorders [14].
Rett syndrome (RTT) is a neurological disorder that almost exclusively affects females with an incidence of 1:10,000 live births [15] due to a loss-of-function mutations of the X-linked methyl-CpG binding protein 2 (MeCP2) gene in approximately 90% of classic RTT cases [16]. RTT subjects develop normally up to 18 months of age after which they undergo a period of neurological regression [15]. RTT affects several organs and systems among which the autonomic nervous system [15], the gastrointestinal tract [17] and the immune system [18] making it eligible as a multisystemic disease [15]. It has been shown that MeCP2 deficiency is able to lead to cytokine dysregulation [18, 19], to influence the expression of FOXP3 [20], an important transcription factor involved in the generation of T-reg cells, and to determine the significant increase of secreted IL-17A [20]. Since the Th17/T-reg balance is implicated in the development of autoimmune/inflammatory disorders it is possible to hypothesize the presence of an autoimmune component in RTT [21]. To this regard, intestinal dysbiosis may cause chronic intestinal inflammation and autoimmunity as occurring in Inflammatory Bowel Diseases (IBDs) [22]. Previous studies indicated the presence of a subclinical inflammatory status in subjects affected by RTT [23] remarked by cytokine dysregulation in both Th1 and Th17 responses [19, 20, 24, 25] and an intestinal dysbiosis characterized by high relative abundance of the genus Candida [26]. Therefore, we asked whether the intestinal fungal population of RTT subjects might be involved in the sub-inflammatory status triggered by MeCP2 deficiency. Here we studied the cultivable gut mycobiota of RTT subjects characterizing the isolated fungi for their virulence-related traits and antifungal resistance. Moreover, we characterized the genetic diversity of C. albicans and C. parapsilosis isolates and their ability to induce innate and adaptive immunological responses in human PBMCs in order to elucidate the role of fungi in RTT gastrointestinal pathophysiology.
Methods
Isolation and identification of cultivable fungal species from faeces
The participants’ data related to the 50 RTT patients and 29 Healthy Controls (HC) included in this study are available in [26]. Stool samples from enrolled subjects [26] were collected, aliquoted as it is and stored at − 80 °C until analysis. Samples were homogenized in sterile Ringer’s solution and plated on solid YPD medium (1% Yeast extract, 2% Bacto-peptone, 2% D-glucose, 2% agar) supplemented with 25 U/ml of penicillin, 25 μg/ml of streptomycin (Sigma-Aldrich) and incubated aerobically at 27 °C for 3–5 days. All fungal isolates grown on the selective medium were further isolated to obtain single-cell pure colonies. Genomic DNA was extracted from pure cultures of the isolated colonies as previously described [27]. Fungal isolates were identified by amplification and sequencing of the ribosomal Internal Transcribed Spacer (ITS) region, using ITS1 (5’-GTTTCCGTAGGTGAACTTGC-3′) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3′) primers [28]. ITS1–4 sequences were then classified by using the BLAST algorithm in the NCBI database (minimum 97% sequence similarity and 95% coverage with a described species).
Invasive growth
The ability of fungal strains to penetrate YPD solid medium was tested as previously described [29]. M28-4D and BY4742 S. cerevisiae strains, known to be invasive and non-invasive respectively, were used as controls. The strain invasiveness was assigned with scores from 3 (highly invasive) to 0 (non-invasive).
Hyphal formation
Fungal cells (~ 105 cells/ml) were grown for 7 days in liquid YPD and YNB media (0.67% Yeast Nitrogen Base w/o aminoacids and (NH4)2SO4 (Sigma-Aldrich), 2% glucose), both at 27 °C and 37 °C in order to evaluate hyphae or pseudohyphae formation. Formation of hyphae was inspected by optical microscope observation with a Leica DM1000 led instrument (magnification 40× and 100×) [30].
Biofilm formation
Biofilm formation was quantified according to a previous published protocol [31]. Briefly, fungal cells (~ 105 cells/ml) were grown in liquid YPD at 37 °C for 48 h in flat-bottom 96-well plates. After the incubation period, cell suspensions were aspirated and each well with the adhered fungal cells was washed three times with deionized H2O and one time with PBS 1X. Biofilm-coated wells were then incubated with 0.01% of crystal violet (Sigma) for 30 min and washed as above. Finally, each well of the dried 96-well plate was incubated with 100 μl of 100% EtOH for 10 min and biofilm formation was quantified by optical density measurement at 570 nm with a microplate reader (Synergy2, BioTek, USA).
Antifungal susceptibility testing
All fungal isolates were tested for susceptibility to fluconazole, itraconazole and 5-flucytosine (Sigma-Aldrich) by Minimum Inibitory Concentration (MIC) assays according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations [32, 33]. EUCAST clinical breakpoints (CBPs) were used to evaluate the antifungal resistance. Although CBPs have not been established for non-Candida yeasts and the non-Aspergillus moulds, they have been used as a proxy for the evaluation of antifungals susceptibility in such isolates.
RAPD genotyping and clustering analysis
Candida albicans and C. parapsilosis isolates were genotyped by Random Amplification of Polymorphic DNA (RAPD) using the primer Oligo 2 (5’-TCACGATGCA-3′) as described previously [34]. Amplifications were performed according to the following protocol: 5 min at 94 °C, 40 cycles of 30s at 94 °C, 30s at 36 °C and 2 min at 72 °C, followed by a final extension of 10 min at 72 °C. The PCR reaction mix contained 1X PCR buffer 2 mM MgCl2, 200 μM of dNTPs, 0.4 μM of the primer, 2.5 U of Taq Polymerase and 10 ng of gDNA as template. PCR amplicons were separated using a 2% agarose gel in 1× TAE buffer at 90 V for 2 h and visualized with 0.5 μg/ml ethidium bromide staining. The presence or absence of an amplicon at any position of the gel was used for the construction of a binary matrix for the calculation of samples’ distance similarity according to the Jaccard index [35] by mean of the “vegdist” function within the vegan R package. The samples have been then clustered hierarchically according to the UPGMA method by using the “hclust” function within the stats R package.
Isolation and stimulation of PBMCs
Human peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque density gradient centrifugation (Biochrom, Berlin, Germany) from buffy coats provided by the Transfusion Unit of Ospedale Santa Chiara in Trento, Italy. The experimental plan was approved by the local hospital ethical committee, and informed consent was obtained from all the healthy donors (protocol No: 54896583). Candida isolates were cultured in YPD medium for 18 h at 37 °C. Fungal cells were harvested by centrifugation, washed twice with PBS, heat-killed for 3 h at 65 °C and resuspended in culture medium (RPMI1640; Sigma Aldrich). For stimulation experiments, 5 × 105 PBMCs in RPMI1640 were incubated with 5 × 106 heat-killed C. albicans, C. parapsilosis or RPMI1640 medium alone (negative control) [36]. After the incubation periods (24 h for IL-1β, IL-6, TNF-α, IL-10 production and 120 h for IL-17A, INFγ, IL-22, IL-10 production) cell suspensions were centrifuged and supernatants were collected and stored at − 20 °C until assayed. Each experiment was performed in triplicate.
Cytokine assays
Cytokine detection i.e. IL-17A, INFγ, IL-22, IL-1β, IL-6, TNF-α, IL-10 production, were assayed using the MAP human cytokine/chemokine kit (Merck Millipore) according to the manufacturer’s instructions (MagPix technology).
Flow cytometry
PBMCs were collected after stimulation with Candida isolates in a ratio of 10:1 (stimuli:cells) and washed with PBS. Intracellular staining for IDO1 (after 24 h of stimulation), T-bet and RORγt (after 5 day of stimulation) were performed using the fixation/permeabilization buffer kit (Life Technologies) following the manufacturing recommendations. Cells were then stained with adequate concentrations of labelled antibodies diluted in PBS with 10% heat-inactivated foetal bovine serum (FBS) for 20 min at room temperature, A minimum of ten thousand events for each sample were acquired using a Guava easyCyte 8 T flow cytometer (Merck Millipore) and analysed using the inCyte software (Merck Millipore). Cells were gated first based on forward and side scatter to exclude dead cells and cell debris. The area of positivity was determined by using an isotype-matched control MAb. Antibodies used: Fluorescein isothiocyanate (FITC)-IDO1 (BD Biosciences Pharmingen, Prodotti Gianni, Italy), FITC-Tbet (Millipore), allophycocyanin (APC)-RORγt (BD Biosciences Pharmingen, Prodotti Gianni, Italy).
Statistical analysis
Wilcoxon rank-sum tests and Spearman’s correlations were performed using the R software [37] through the stats R package (version 3.1.2) and the psych R package, respectively. Permutational MANOVA (PERMANOVA) test was performed using the adonis()function of the vegan R package with 999 permutations. All p-values have been corrected for multiple hypothesis controlling the false discovery rate (FDR) [38].
Results
RTT gut mycobiota shows a reduction of C. albicans and an increase of C. parapsilosis populations
We identified 122 fungal isolates belonging to different species (Additional file 1: Table S1). Twenty-four of such isolates were obtained from stool samples of RTT subjects (Additional file 1: Table S1). We discovered a significant reduction of fungal species richness in RTT subjects compared to HC (p = 3.9e-05, Wilcoxon rank-sum test) in agreement with the results obtained using a metagenomic approach from the same study cohort [26]. Candida was the most abundant genus present in both RTT subjects (91.7%) and HC (71.4%) with C. albicans and C. parapsilosis as the two most abundant species in both RTT subjects and HC. Interestingly, we observed an inversion in the relative abundances of C. albicans and C. parapsilosis between RTT subjects and HC. While in RTT subjects 4 out of 24 fungal isolates belonged to C. albicans (16.7%) and 14 out of 24 belonged to C. parapsilosis (58.3%), in the HC 49 out of 98 fungal isolates belonged to C. albicans (50%) and 15 out of 98 belonged to C. parapsilosis (15.3%) (Additional file 2: Figure S1). We then characterized the fungal isolates for putative virulence-associated traits and resistance to antifungals (Additional file 1 Table S1). We found that 50% and 63.8% of fungal isolates from RTT subjects and HC, respectively, were able to form hyphae or pseudohyphae (Additional file 1: Table S1). In addition, we observed that the morphotype switch to hyphae or pseudohyphae was related to the isolates’ invasiveness, with hyphae- and pseudohyphae-forming isolates being the most invasive (Additional file 3: Figure S2A). We also observed that RTT isolates produced more biofilm (p = 1.3e-05, Wilcoxon rank-sum test; Additional file 3: Figure S2B) and were significantly more resistant to fluconazole compared to HC isolates (45.8% of RTT isolates were resistant vs 18.1% of HC isolates; p = 5.1e-06, Wilcoxon rank-sum test). As previously observed, we found the co-occurrence of azole cross-resistance between fluconazole and itraconazole (Spearman’s correlation r = 0.57; p = 2.2e-10) [39]. Almost the totality of the isolates (96.7%) were susceptible to 5-flucytosine, with MIC≤0.125 μg/ml (Additional file 1: Table S1). Candida parapsilosis isolates from HC were susceptible to fluconazole (MIC90 = 2 μg/ml; R = 7.1%) and itraconazole (MIC90 = 0.0156 μg/ml; R = 0%) while C. parapsilosis isolates from RTT subjects exhibited a high resistance to these antifungals (fluconazole, MIC90 > 64 μg/ml, R = 35.7%, p = 0.003, Wilcoxon rank-sum test, Figure 1A; itraconazole, MIC90 > 8 μg/ml, R = 35.7%; Table 1). On the contrary, C. albicans isolates from HC were resistant to fluconazole (MIC90 > 64 μg/ml, R = 24.4%) and itraconazole (MIC90 > 8 μg/ml, R = 63.4%; p = 0.03, Wilcoxon rank-sum test; Fig. 1B) while only one RTT C. albicans isolate was resistant to fluconazole (Table 1). Noteworthy, all other Candida species isolated from RTT subjects (i.e. C. glabrata, C. pararugosa and C. tropicalis) were resistant to fluconazole (MIC90 > 64 μg/ml; R = 100%) and itraconazole (MIC90 = 8 μg/ml; R = 100%) while Candida spp. isolated from HC (i.e. C. deformans, C. intermedia and C. lusitaniae) were completely susceptible to these azoles (Table 1). Taken together these results suggest that RTT isolates may be more difficult to eradicate in case of infection than HC isolates. Finally, we have been able to isolate Trichosporon asteroides and Saccharomyces cerevisiae only from RTT subjects. These isolates were both resistant to fluconazole (with MIC>64 μg/ml and MIC = 8 μg/ml respectively) while Trichosporon asteroides was also resistant to itraconazole (MIC>8 μg/ml). Such species are recognized as potential new emerging fungal pathogens [40] thus representing a potential threat for RTT subjects.
C. parapsilosis isolates from RTT subjects and HC are genetically distinct
The genetic diversity among Candida isolates was determined by UPGMA hierarchical clustering analysis of Jaccard distances calculated from RAPD genotyping. We observed that C. parapsilosis isolates from RTT samples were genetically unrelated to those from HC, with most of RTT C. parapsilosis isolates clustering in a single group (Fig. 2 and Additional file 4: Figure S3; p = 0.002, PERMANOVA). On the contrary, C. albicans isolates from RTT subjects were genetically more diverse, clustering in different clades of the tree (Additional file 5: Figure S4; p = 0.779, PERMANOVA). It is worth noting that we only obtained 4 C. albicans isolates from RTT samples.
Candida parapsilosis from RTT subjects induces high levels of IL-10 in PBMCs
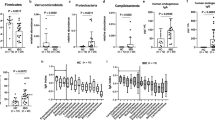
The first step in the immunological response against Candida is the production by innate immune cells of pro-inflammatory cytokines, such as IL-1β, IL-6 and TNFα. Successively, these cytokines promote adaptive immunity mediated by Th1 or Th17 responses [2]. Stimulation of PBMCs with C. albicans and C. parapsilosis isolates from HC and RTT subjects revealed that RTT Candida parapsilosis isolates induced higher levels of IL-1β (p = 0.01, Wilcoxon rank-sum test) and, although only close to statistical significance, TNFα (p = 0.058, Wilcoxon rank-sum test) with respect to HC Candida parapsilosis isolates (Fig. 3). On the contrary, no significant differences were observed in the levels of IL-17A, IL-22 and INFγ (Fig. 3). Furthermore we observed that C. parapsilosis isolates from RTT subjects induced highly significant levels of IL-10 compared to HC C. parapsilosis isolates and RTT C. albicans isolates (p < 0.003, Wilcoxon rank-sum test; Fig. 3) suggesting an increased fungal tolerance towards these C. parapsilosis isolates, potentially favouring fungal persistence within the host. Nevertheless, we did not observe significant differences in the expression of IDO1 in PBMCs stimulated by C. parapsilosis isolates (median HC = 30%, IQR = 22.8–39.3%; median RTT = 44%, IQR = 29.1–48.3%; p = 0.24, Wilcoxon rank-sum test). Since we observed variable levels of Th-driving cytokines, we asked whether Candida isolates were able to induce a different Th1/Th17 polarization. Therefore, we measured the intracellular levels of the key transcription factors T-bet and RORγt (Additional file 6: Figure S5), involved in the differentiation of CD4+ naïve cells in Th1 and Th17 cell, respectively [41]. As previously observed in culture supernatants, we measured variable, but not statistically significant levels of T-bet and RORγt in response to the different C. parapsilosis and C. albicans isolates (Additional file 6: Figure S5), reflecting the potential of different strains to elicit an immune reaction at different extents. This could be due to the diverse immune reactivity shown by the different isolates of the same species, as previously reported [42, 43]. However, C. parapsilosis isolates from RTT subjects induced more CD4+ cells co-expressing both RORγt and T-bet compared to HC C. parapsilosis isolates (raw p-value = 0.04, FDR-corrected p-value = 0.12; Wilcoxon rank-sum test; Additional file 6: Figure S5c).
Cytokines production by peripheral blood mononuclear cells (PBMCs; 5 × 105 cell) after stimulation with 5 × 106 heat-killed C. albicans or C. parapsilosis isolates from HC and RTT subjects. In panels a-d) are reported the values for the cytokines produced by innate immune cells (IL-1β, IL-6, TNFα and IL-10 after 24 h of PBMCs stimulation) while in e-h) the values for the cytokines produced following adaptive immune responses (IL-17A, IL-22, IFNγ and IL-10 after 5 days of PBMCs stimulation). The dots represent each of the three replicates per isolate tested; *p < 0.05, Wilcoxon rank-sum test
Discussion
In the present study we show that in RTT, a multisystemic neurological disorder, faecal C. parapsilosis isolates hold phenotypic traits potentially favouring the previously observed low-grade intestinal inflammatory status [26]. Species-level analysis of the cultivable gut mycobiota revealed C. parapsilosis as the most abundant yeast species in RTT subjects, genetically distinct from HC C. parapsilosis isolates. Interestingly, RTT C. parapsilosis isolates were characterized by high levels of resistance to azoles antifungals. Furthermore, the high levels of IL-10 produced by PBMCs in response to RTT C. parapsilosis isolates suggest that these isolates have developed the capacity to persist within the host. IL-10 usually exert a homeostatic control to keep inflammation under control, although high levels of IL-10 are characteristic of chronic fungal infections dominated by non-resolving inflammation [2]. It has been observed that C. albicans induces host’s immunosuppression by increased IL-10 production by immune cells representing an important mechanisms in Candida pathogenesis [44]. Likewise, RTT C. parapsilosis isolates could escape immune clearance through a mechanism mediated by high levels of IL-10 that, in turn, could impair antifungal Th1 immunity, thus favouring a persistent intestinal colonization.
C. parapsilosis has been described as one of the leading causes of invasive candidiasis [45], being responsible of macrophage activation and allergic airways inflammation [46] and to be one of the dominant Candida species leading to dysbiosis in IBDs [47]. Fungal opportunistic infections are generally ascribed to defective host immunity, although they could require specific microbial population dysbiosis [48], as recently observed in RTT [26]. Recent studies indicated that fungal infections may originate from individual’s own commensal strains and that the ability of a commensal organism to produce disease is not merely a consequence of impaired host immunity [49]. Indeed, C. albicans passage through the GI tract results in a phenotypic switch in the so-called gastrointestinal induced transition cells, where virulence-associated genes are down-regulated enabling fungal adaptation for long-term survival in the large intestine [50]. Recent studies further show how strain specific differences in morphogenetic networks, regulating the switch from hyphal and yeast forms, subtend differences in their pathogenic potential, thus suggesting the importance to move the metagenomics analyses to the strain level [51]. Therefore, C. parapsilosis isolates from RTT subjects may be commensals potentially hazardous for the host due not only to RTT’s altered immunological status [19] but also by the presence of a dysbiotic gut microbiota [26]. Moreover, RTT C. parapsilosis isolates induced a higher proportion of a mixed Th1/Th17 cells population compared to HC C. parapsilosis isolates. Although Th1 and Th17 responses usually counter-regulate each other, there are increasing evidences of co-operation and dependency between these two immunological responses [52] which are involved in chronic, pro-inflammatory responses as observed in IBDs [53] potentially resulting in adaptive immunity against the commensal microbiota [54]. Interestingly, it has been previously shown that MeCP2 could actually play a regulatory role in T-cell resilience to inflammation [20]. Emerging evidence indicates that MeCP2 deficiency is able to lead to cytokine dysregulation including macrophage-related cytokines in Mecp2-null mice and RTT females [18, 19], although the understanding of the molecular mechanisms underlying this pro-inflammatory status remains elusive.
We have previously evidenced an inflammatory process in both RTT patients [19, 23] and animal models [55]. Biochemical analyses showed significant changes in the expression of acute phase response (APR) and immune system proteins in the serum of RTT subjects [23]. A direct relationship between MeCP2 and the immune system has been demonstrated, since MeCP2 is important for the differentiation of naïve CD4+ T cells into Th17 cells and for the commitment of naïve CD4+ T cells to the Th1 lineage [24]. Indeed, MeCP2 plays a critical role in promoting multiple cytokine-mediated signalling pathways through the MeCP2-miR-124-SOCS5 axis. This signalling pathway is required for the activation of signal transducer and activator of transcription 3 (STAT3) and STAT1 in CD4+ T cells, with consequent generation of Th17 cells [24]. The mechanisms underlying the inflammatory process appear to be related to a severe cytokine dysregulation, possibly reflecting a macrophage dysregulation/dysfunction, as previously suggested for Mecp2−/− mice [56]. Furthermore, there is a critical interplay between inflammation and oxidative stress in the underlying mechanisms [57]. More recently, MeCP2 has been reported to act as an epigenetic regulator of immune and inflammatory responses during zebrafish development [58]. In this context, the bacterial and fungal microbiota dysbiosis demonstrated in our previous metataxonomics study from the same cohort of subjects [26] and the presence of putative virulent, pro-inflammatory intestinal C. parapsilosis strains could represent an additional factor in RTT’s gastrointestinal pathophysiology and subclinical inflammation.
Conclusions
The gut mycobiota is emerging as a key player in maintaining the overall microbial community structure of the human gut, and main actor in host physiopathology [59]. Previous observations from our team indicated the presence of a subclinical inflammatory status [23], cytokine dysregulation [19] and intestinal dysbiosis [26] contributing to gastrointestinal symptoms in Rett syndrome. In the present manuscript, we moved our analysis to the strain level, investigating whether fungal isolates from RTT subjects may favour the sub-inflammatory status triggered by MeCP2 deficiency. Our results discovered C. parapsilosis, but not C. albicans, as the most abundant and potentially pro-inflammatory yeast species in the gut of RTT subjects. We propose that intestinal dysbiosis and the presence of pro-inflammatory C. parapsilosis strains could have a role in RTT’s gastrointestinal abnormalities laying the basis for the design of novel therapeutic strategies that, by targeting specific components of the gut microbiota, would restore eubiosis and intestinal physiology in RTT.
Abbreviations
- CBP:
-
Clinical breakpoint
- HC:
-
Healthy controls
- IBDs:
-
Inflammatory bowel diseases
- IDO1:
-
Indoleamine 2,3-dioxygenase 1
- IL:
-
Interleukin
- INF:
-
Interferon, IQR, interquartile range
- MeCP2:
-
Methyl-CpG binding protein 2
- MIC:
-
Minimum inhibitory concentration
- PBMC:
-
Peripheral blood mononuclear cells
- RAPD:
-
Random amplification of polymorphic DNA
- RTT:
-
Rett syndrome
- Th:
-
T-helper
- TNF:
-
Tumor necrosis factor
- T-reg cell:
-
Regulatory T cell
- UPGMA:
-
Unweighted pair group method with arithmetic mean
References
Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14(6):405–16.
Romani L. Immunity to fungal infections. Nat Rev Immunol. 2011;11(4):275–88.
Vinh DC, Sugui JA, Hsu AP, Freeman AF, Holland SM. Invasive fungal disease in autosomal-dominant hyper-IgE syndrome. J Allergy Clin Immunol. 2010;125(6):1389–90.
Puel A, Picard C, Cypowyj S, Lilic D, Abel L, Casanova JL. Inborn errors of mucocutaneous immunity to Candida albicans in humans: a role for IL-17 cytokines? Curr Opin Immunol. 2010;22(4):467–74.
Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4(10):762–74.
Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478(7368):197–203.
Fallarino F, Grohmann U, You S, McGrath BC, Cavener DR, Vacca C, Orabona C, Bianchi R, Belladonna ML, Volpi C, et al. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176(11):6752–61.
Romani L, Zelante T, De Luca A, Iannitti RG, Moretti S, Bartoli A, Aversa F, Puccetti P. Microbiota control of a tryptophan-AhR pathway in disease tolerance to fungi. Eur J Immunol. 2014;44(11):3192–200.
Neville BA, d'Enfert C, Bougnoux ME. Candida albicans commensalism in the gastrointestinal tract. FEMS Yeast Res. 2015;15(7):fov081.
Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–35.
De Luca A, Montagnoli C, Zelante T, Bonifazi P, Bozza S, Moretti S, D'Angelo C, Vacca C, Boon L, Bistoni F, et al. Functional yet balanced reactivity to Candida albicans requires TRIF, MyD88, and IDO-dependent inhibition of Rorc. J Immunol. 2007;179(9):5999–6008.
Cheng SC, van de Veerdonk F, Smeekens S, Joosten LA, van der Meer JW, Kullberg BJ, Netea MG. Candida albicans dampens host defense by downregulating IL-17 production. J Immunol. 2010;185(4):2450–7.
Bozza S, Fallarino F, Pitzurra L, Zelante T, Montagnoli C, Bellocchio S, Mosci P, Vacca C, Puccetti P, Romani L. A crucial role for tryptophan catabolism at the host/Candida albicans interface. J Immunol. 2005;174(5):2910–8.
Schwartz CE. Aberrant tryptophan metabolism: the unifying biochemical basis for autism spectrum disorders? Biomark Med. 2014;8(3):313–5.
Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56(3):422–37.
Liyanage VR, Rastegar M. Rett syndrome and MeCP2. NeuroMolecular Med. 2014;16(2):231–64.
Motil KJ, Caeg E, Barrish JO, Geerts S, Lane JB, Percy AK, Annese F, McNair L, Skinner SA, Lee HS, et al. Gastrointestinal and nutritional problems occur frequently throughout life in girls and women with Rett syndrome. J Pediatr Gastroenterol Nutr. 2012;55(3):292–8.
Cronk JC, Derecki NC, Litvak V, Kipnis J. Unexpected cellular players in Rett syndrome pathology. Neurobiol Dis. 2016;92(Pt A):64–71.
Leoncini S, De Felice C, Signorini C, Zollo G, Cortelazzo A, Durand T, Galano JM, Guerranti R, Rossi M, Ciccoli L, et al. Cytokine Dysregulation in MECP2- and CDKL5-Related Rett Syndrome: Relationships with Aberrant Redox Homeostasis, Inflammation, and omega-3 PUFAs. Oxidative Med Cell Longev. 2015;2015:421624.
Li C, Jiang S, Liu S-Q, Lykken E, Zhao L-T, Sevilla J, Zhu B, Li Q-J. MeCP2 enforces Foxp3 expression to promote regulatory T cells’ resilience to inflammation. Proc Natl Acad Sci. 2014;111(27):E2807–16.
De Felice C, Leoncini S, Signorini C, Cortelazzo A, Rovero P, Durand T, Ciccoli L, Papini AM, Hayek J. Rett syndrome: An autoimmune disease? Autoimmun Rev. 2016;15(4):411–6.
Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–23.
Cortelazzo A, De Felice C, Guerranti R, Signorini C, Leoncini S, Pecorelli A, Zollo G, Landi C, Valacchi G, Ciccoli L, et al. Subclinical inflammatory status in Rett syndrome. Mediat Inflamm. 2014;2014:480980.
Jiang S, Li C, McRae G, Lykken E, Sevilla J, Liu SQ, Wan Y, Li QJ. MeCP2 reinforces STAT3 signaling and the generation of effector CD4+ T cells by promoting miR-124-mediated suppression of SOCS5. Sci Signal. 2014;7(316):ra25.
O'Driscoll CM, Lima MP, Kaufmann WE, Bressler JP. Methyl CpG binding protein 2 deficiency enhances expression of inflammatory cytokines by sustaining NF-kappaB signaling in myeloid derived cells. J Neuroimmunol. 2015;283:23–9.
Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, Jousson O, Leoncini S, Pindo M, Renzi D, et al. Altered gut microbiota in Rett syndrome. Microbiome. 2016;4(1):41.
Hoffman CS, Winston F. A 10-Minute DNA Preparation from Yeast Efficiently Releases Autonomous Plasmids for Transformation of Escherichia-Coli. Gene. 1987;57(2–3):267–72.
Sebastiani F, Barberio C, Casalone E, Cavalieri D, Polsinelli M. Crosses between Saccharomyces cerevisiae and Saccharomyces bayanus generate fertile hybrids. Res Microbiol. 2002;153(1):53–8.
Vopalenska I, Hulkova M, Janderova B, Palkova Z. The morphology of Saccharomyces cerevisiae colonies is affected by cell adhesion and the budding pattern. Res Microbiol. 2005;156(9):921–31.
Strati F, Di Paola M, Stefanini I, Albanese D, Rizzetto L, Lionetti P, Calabro A, Jousson O, Donati C, Cavalieri D, et al. Age and Gender Affect the Composition of Fungal Population of the Human Gastrointestinal Tract. Front Microbiol. 2016;7:1227.
Jin Y, Yip HK, Samaranayake YH, Yau JY, Samaranayake LP. Biofilm-forming ability of Candida albicans is unlikely to contribute to high levels of oral yeast carriage in cases of human immunodeficiency virus infection. J Clin Microbiol. 2003;41(7):2961–7.
Rodriguez-Tudela J, Arendrup M, Barchiesi F, Bille J, Chryssanthou E, Cuenca-Estrella M, Dannaoui E, Denning D, Donnelly J, Dromer F. EUCAST Definitive Document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts: Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST)∗. Clin Microbiol Infect. 2008;14(4):398–405.
Rodriguez-Tudela J, Arendrup M, Arikan S, Barchiesi F, Bille J, Chryssanthou E, Cuenca-Estrella M, Dannaoui E, Denning D, Donnelly J. EUCAST Definitive document E. DEF 9.1: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia forming moulds. Def. 2008;9:1–13.
Binelli CA, Moretti ML, Assis RS, Sauaia N, Menezes PR, Ribeiro E, Geiger DC, Mikami Y, Miyaji M, Oliveira MS, et al. Investigation of the possible association between nosocomial candiduria and candidaemia. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2006;12(6):538–43.
Levandowsky M, Winter D. Distance between sets. Nature. 1971;234(5323):34–5.
Toth A, Csonka K, Jacobs C, Vagvolgyi C, Nosanchuk JD, Netea MG, Gacser A. Candida albicans and Candida parapsilosis induce different T-cell responses in human peripheral blood mononuclear cells. J Infect Dis. 2013;208(4):690–8.
Team RC. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012. ISBN 3-900051-07-0; 2014
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300.
Pfaller MA, Diekema DJ. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J Clin Microbiol. 2012;50(9):2846–56.
Cornely OA, Cuenca-Estrella M, Meis JF, Ullmann AJ. European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Fungal Infection Study Group (EFISG) and European Confederation of Medical Mycology (ECMM) 2013 joint guidelines on diagnosis and management of rare and emerging fungal diseases. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2014;20(Suppl 3):1–4.
Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. 2010;28:445–89.
Rizzetto L, Giovannini G, Bromley M, Bowyer P, Romani L, Cavalieri D. Strain dependent variation of immune responses to A. fumigatus: definition of pathogenic species. PLoS One. 2013;8(2):e56651.
Marakalala MJ, Vautier S, Potrykus J, Walker LA, Shepardson KM, Hopke A, Mora-Montes HM, Kerrigan A, Netea MG, Murray GI, et al. Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. PLoS Pathog. 2013;9(4):e1003315.
Netea MG, Sutmuller R, Hermann C, Van der Graaf CA, Van der Meer JW, van Krieken JH, Hartung T, Adema G, Kullberg BJ. Toll-like receptor 2 suppresses immunity against Candida albicans through induction of IL-10 and regulatory T cells. J Immunol. 2004;172(6):3712–8.
Trofa D, Gacser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev. 2008;21(4):606–25.
Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Nunez G, Shibuya A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE(2). Cell Host Microbe. 2014;15(1):95–102.
Ott SJ, Kuhbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol. 2008;43(7):831–41.
Casadevall A, Pirofski LA. Accidental virulence, cryptic pathogenesis, martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryot Cell. 2007;6(12):2169–74.
Odds FC, Davidson AD, Jacobsen MD, Tavanti A, Whyte JA, Kibbler CC, Ellis DH, Maiden MC, Shaw DJ, Gow NA. Candida albicans strain maintenance, replacement, and microvariation demonstrated by multilocus sequence typing. J Clin Microbiol. 2006;44(10):3647–58.
Pande K, Chen C, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013;45(9):1088–91.
Cavalieri D, Di Paola M, Rizzetto L, Tocci N, De Filippo C, Lionetti P, Ardizzoni A, Colombari B, Paulone S, Gut IG, et al. Genomic and Phenotypic Variation in Morphogenetic Networks of Two Candida albicans Isolates Subtends Their Different Pathogenic Potential. Front Immunol. 2017;8:1997.
Damsker JM, Hansen AM, Caspi RR. Th1 and Th17 cells: adversaries and collaborators. Ann N Y Acad Sci. 2010;1183:211–21.
Strober W, Fuss IJ. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140(6):1756–67.
Jyonouchi H, Geng L, Cushing-Ruby A, Monteiro IM. Aberrant responses to TLR agonists in pediatric IBD patients; the possible association with increased production of Th1/Th17 cytokines in response to candida, a luminal antigen. Pediatric allergy and immunology: official publication of the European Society of Pediatric Allergy and Immunology. 2010;21(4 Pt 2):e747–55.
Cortelazzo A, De Felice C, De Filippis B, Ricceri L, Laviola G, Leoncini S, Signorini C, Pescaglini M, Guerranti R, Timperio AM, et al. Persistent Unresolved Inflammation in the Mecp2-308 Female Mutated Mouse Model of Rett Syndrome. Mediat Inflamm. 2017;2017:9467819.
Cronk JC, Derecki NC, Ji E, Xu Y, Lampano AE, Smirnov I, Baker W, Norris GT, Marin I, Coddington N, et al. Methyl-CpG Binding Protein 2 Regulates Microglia and Macrophage Gene Expression in Response to Inflammatory Stimuli. Immunity. 2015;42(4):679–91.
De Felice C, Signorini C, Leoncini S, Durand T, Ciccoli L, Hayek J. Oxidative stress: a hallmark of Rett syndrome. Future Neurol. 2015;10(3):179–82.
van der Vaart M, Svoboda O, Weijts BG, Espin-Palazon R, Sapp V, Pietri T, Bagnat M, Muotri AR, Traver D. Mecp2 regulates tnfa during zebrafish embryonic development and acute inflammation. Dis Model Mech. 2017;10(12):1439–51.
Iliev ID, Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol. 2017;17(10):635–46.
Acknowledgements
The authors would like to thank Irene Stefanini and Davide Albanese for the helpful discussion. We thank also Unifarm S.p.A. for the financial support to FS.
Funding
This work was financially supported from the “Accordo di Programma Integrato MetaFoodLabs” funded by the research office of the Provincia Autonoma di Trento (Italy) (PAT Prot. S116/2012/537723) and by the EU Framework Programme 7 Collaborative Project [242220]-JPI ENPADASI. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article (and its supplementary information files).
Author information
Authors and Affiliations
Contributions
FS designed and performed the experiments, analysed the data and wrote the manuscript. LR supervised and contributed to the immunological assays. CD supervised and contributed to data analysis. SL, CDFe, JH, DR and AC recruited subjects and collected specimens. CD, DC and CDF provided the reagents for the study. DC and CDF conceived the study and approved the final manuscript. All the authors critically reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the institutional review board of the Siena University Hospital (AOUS, Siena, Italy) and all enrolled subjects or tutors gave written informed consent in accordance with the sampling protocol approved by the local Ethical Committee (No: 2012–005021-76). The in vitro study using human cells was designed in conformity with the international recommendation (Dir. EU 2001/20/EC) and its Italian counterpart (DM 15 Luglio 1997; D.Lvo 211/2003; D.L.vo 200/2007) for clinical trial and following the Declaration of Helsinki, to assure protection and care of subjects involved. The study was approved by the Ethical Committee of Ospedale Santa Chiara di Trento (nr. 54,896,583).
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Phenotypic characteristics and antifungals susceptibility of fungal isolates. §, MIC ranges: fluconazole 0.125–64 μg/ml; itraconazole 0.0156–8 μg/ml; 5-flucytosine 0.125–64 μg/ml; #, 0 = non-invasive; 1 = poor invasive; 2 = invasive; 3 = very invasive. * measured by optical density at 570 nm; NA, not applicable. (DOCX 39 kb)
Additional file 2:
Figure S1. Relative abundances of Candida albicans and Candida parapsilosis isolates in Rett syndrome subjects (RTT) and healthy controls (HC). The total abundance of all the other fungal isolates is also reported as “other species” (PDF 22 kb)
Additional file 3:
Figure S2. a) Intestinal fungal isolates ability (or not) to produce hyphae or pseudo-hyphae in relationship with their ability to be invasive on YPD solid medium; b) biofilm production by intestinal fungal isolates from HC and RTT subjects; ***p < 0.0001, Wilcoxon rank-sum test. (PDF 35 kb)
Additional file 4:
Figure S3. UPGMA hierarchical clustering of C. parapsilosis genetic diversity calculated by using samples’ distance similarities (Jaccard index) from RAPD genotyping. C. parapsilosis isolates from HC and RTT subjects in green and red, respectively. (PDF 15 kb)
Additional file 5:
Figure S4. UPGMA hierarchical clustering of C. albicans genetic diversity calculated by using samples’ distance similarities (Jaccard index) from RAPD genotyping. C. albicans isolates from HC and RTT subjects in green and red, respectively; in gray the lab strain SC5314. (PDF 16 kb)
Additional file 6:
Figure S5. Percentage of positive T-cells to T-bet, RORγt and both transcription factors T-bet and RORγt, as measured by intracellular staining and flow cytometry of PBMCs stimulated with a, b, c) C. parapsilosis isolates and d, e, f) C. albicans isolates from HC and RTT subjects. Cells were gated for CD4+ and data are given as percentage of total gated CD4+ cells. (PDF 41 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Strati, F., Calabrò, A., Donati, C. et al. Intestinal Candida parapsilosis isolates from Rett syndrome subjects bear potential virulent traits and capacity to persist within the host. BMC Gastroenterol 18, 57 (2018). https://doi.org/10.1186/s12876-018-0785-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12876-018-0785-z