Abstract
Background
Inflammatory bowel disease (IBD) is associated with increased intestinal permeability, which involves paracellular passage regulated through tight junctions (TJ). The aim of the study was to investigate single nucleotide polymorphisms (SNP) located in genes encoding interacting TJ proteins and corresponding expressions, in relation to IBD.
Methods
Allelic associations between TJ-related genes (F11R, MAGI1, MAGI2, MAGI3, PARD3, PTEN, and TJP1) and IBD, Crohn’s disease (CD), or ulcerative colitis (UC) were investigated. PTPN22 was included since it’s located in the same genetic region as MAGI3. Gene expression levels were investigated in relation to genotype, inflammatory status, phenotype, and medical treatment.
Results
The two strongest allelic associations were observed between IBD and SNPs in MAGI2 (rs6962966) and MAGI3 (rs1343126). Another MAGI3 SNP marker (rs6689879) contributed to increased ileal MAGI3 expression level in non-IBD controls. Furthermore, association between inflammation and decreased expression levels of MAGI3, PTEN, and TJP1 in colonic IBD as well as UC mucosa, and between inflammation and increased expression of PTPN22 in colonic IBD mucosa, was observed.
Conclusions
Our findings lend support to a genetic basis for modulation of intestinal epithelial barrier in IBD, and we have identified MAGI3 as a new candidate gene for IBD.
Similar content being viewed by others
Background
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), are complex diseases thought to result from loss of homeostasis between the intestinal microbial milieu, the immune system, and a genetic predisposition [1]. Genetic association in IBD has been the focus of much research [2, 3] leading to the recent identification of a large number of susceptibility loci [3,4,5]. Still, the hitherto identified genetic associations only account for a small fraction of the heritability of CD and UC [3].
IBD has been linked to increased paracellular permeability [6, 7]. The intestinal permeability is further increased in both CD patients and their relatives, indicating underlying hereditary factors [8]. However, it remains controversial whether this affected permeability is primary, caused by genetic factors, and/or secondary to inflammation or environmental factors. The tight junction (TJ) structure is critical for the permeability properties of the intestine [9] and several studies provide support for a genetic basis in increased permeability [10].
Wapenaar et al. [11] have described an association between celiac disease and single nucleotide polymorphism (SNP) markers in MAGI2 and PARD3. This MAGI2 marker was also associated with UC [11]. Additionally, McGovern et al. [12] identified a link between genetic variation in MAGI2 and both CD and UC.
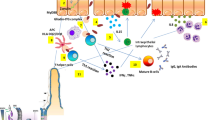
The aim of this study was to investigate relations between IBD and MAGI2 and PARD3, as well as other TJ genes (F11R, MAGI1, MAGI3, PTEN, and TJP1) encoding products interacting with each other (Fig. 1) [13,14,15]. Additionally, PTPN22 was included since it is located in the same genetic region as MAGI3 (Fig. 2) and has previously been described in relation to IBD [5, 16]. To gain a better understanding of the pathogenic mechanism of IBD we further analyzed ileal and colonic gene expression in relation to genotype, inflammatory status, phenotype, and ongoing medical treatment.
Illustration of the genetic region of MAGI3 and PTPN22. SNP data were obtained from the HapMap database (available at: www.hapmap.org; HapMap Data Rel 28 Phase II + III, August10) and analyzed using Haploview version 4.2 (available at www.broad.mit.edu/haploview) with blocks defined according to Gabriel et al. [17]
Methods
Study subjects
Genetic association was investigated in adult Swedish patients with IBD (138 and 157 patients with CD or UC, respectively) and controls from an anonymized regional DNA bank consisting of randomly selected individuals (n = 423) living in the southeastern part of Sweden was used after verbal permission from Prof. Peter Söderkvist (Department of Clinical and Experimental Medicine, Linköping University, Linköping, Sweden) (Table 1, subgroup 1).
A second Swedish cohort (Table 2, subgroup 2), from which both DNA and RNA were available, was recruited to follow up the case–control study of subgroup 1. Blood samples and intestinal biopsy specimens were obtained from adult IBD patients and non-IBD controls. Each intestinal biopsy was categorized as inflamed or non-inflamed based on a compound evaluation of endoscopic findings assessed by one experienced endoscopist (S.A.) and routine histopathologic assessment for inflammation. Only biopsies with concordant results were analyzed further. In total the study included biopsies from 52 Swedish IBD patients (42 inflamed biopsies and 55 non-inflamed biopsies), including 21 CD patients (16 inflamed biopsies and 24 non-inflamed biopsies), 29 UC patients (24 inflamed biopsies and 29 non-inflamed biopsies), 2 IBD-type unclassified (IBDU; 2 inflamed biopsies and 2 non-inflamed biopsies), and 33 non-inflamed non-IBD controls (86 biopsies).
SNP selection for genetic association studies
TJ-related genes (F11R, MAGI1, MAGI2, MAGI3, PARD3, PTEN, and TJP1) encoding products interacting with each other [13, 14] were investigated (Fig. 1). Additionally, PTPN22 was included since it is located in the same genetic region as MAGI3 (Fig. 2) and has previously been described in relation to IBD [5, 16]. All SNP markers are given in Additional file 1: Table S1.
SNP markers (minor allele frequency ≥10%, pair-wise r 2 ≥ 0.8) of F11R, MAGI1, MAGI3, and PTEN were selected using SNPbrowser Software version 4.0 (Applied Biosystems, Foster City, CA). SNP markers for MAGI1 and MAGI3 were limited to exons and exon-intron boundaries, due to the large size of these genes. All SNP markers in the genetic region of MAGI2 [11, 12], MAGI3-PTPN22 [5], PARD3 [11], TJP1 [11], and an additional marker for MAGI1 (rs9880851) [11] were selected from the literature.
Definition of genetic blocks of MAGI3 and PTPN22
Blocks of genetic linkage disequilibrium in the region of MAGI3 and PTPN22 were identified using data from HapMap (available at: www.hapmap.org; HapMap Data Rel. 28 Phase II + III, August10) (Fig. 2). The blocks were visualized using Haploview and the algorithm by Gabriel et al. [17] based on 95% confidence bounds on D’ (Haploview version 4.2, available at: www.broad.mit.edu/haploview).
Genotyping
DNA was isolated from buffy coat or whole blood (EDTA blood) using the MagNA Pure LC DNA Isolation Kit and MagNA Pure extraction robot (Roche, Basel, Switzerland). In patients where no DNA from blood was available, the genotyping was performed using DNA isolated from intestinal biopsies (isolation previously described [18]). Allelic discrimination was carried out using either the TaqMan OpenArray system or real-time PCR, as well as TaqMan SNP genotyping assays (Applied Biosystems, Foster City, CA) (Additional file 1: Table S1).
Allelic discrimination using TaqMan OpenArray Genotyping Plates, TaqMan OpenArray Genotyping Master Mix, GeneAmp PCR System 9700, and OpenArray NT Imager (Applied Biosystems) was in accordance with the manufacturer’s recommendation. Genotype data were analyzed using OpenArray SNP Genotyping Analysis Software version 1.0.3 and TaqMan Genotyper Software v.1.3 (Applied Biosystems).
The allelic discrimination using real-time PCR was performed in a total reaction volume of 10 μL, consisting of TaqMan SNP genotyping assays and TaqMan Genotyping Master Mix using either the 7500 Fast Real-Time PCR System (Applied Biosystems) or the CFX96 Real-time System, C1000 Touch Thermal Cycler (Bio-Rad Laboratories Inc., Hercules, CA). The genotype data was analyzed using 7500 Software version 2.0.6 (Applied Biosystems) or Bio-Rad CFX Manager 3.1 (Bio-Rad Laboratories Inc.).
Gene expression analysis
For genes with a significant genetic association to IBD, CD, or UC (in this study), gene expression was analyzed in relation to genotype, inflammatory status, phenotype, and ongoing medical treatment.
RNA purification was performed as described previously [18]. cDNA was synthesized from 2 μg RNA in a total volume of 40 μL using High Capacity cDNA Reverse Transcription kit with RNase Inhibitor according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). The cDNA was diluted to 25 ng/μL using 0.1 × TE buffer and stored at −80 °C until analysis.
Gene expression of F11R (Hs00170991_m1), MAGI2 (Hs00202321_m1), MAGI3 (Hs00326365_m1), PTEN (Hs02621230_s1), PTPN22 (Hs01587518_m1), and TJP1 (Hs01551861_m1) was analyzed using TaqMan Gene Expression Assay (Applied Biosystems), TaqMan Universal Mastermix (Applied Biosystems), and a 7500 Fast Real-Time PCR System (Applied Biosystems). Each individual reaction contained 10 ng cDNA in a total reaction volume of 20 μL.
Threshold cycle (CT) values were established (ExpressionSuite Software Version 1.0.3; Applied Biosystems) and normalized to the average of selected reference genes (CASC3 (Hs00201226_m1), UBA52 (Hs03004332_g1), and POP4 (Hs00198357_m1) [18]) generating delta-CT (ΔCT) values. Relative quantification (RQ) values were further established, according to Livak et al. [19], by relating the ΔCT value to the sample with the lowest gene expression for each gene (using Microsoft Office Excel; Microsoft Corporation, Redmond, WA). Group differences in gene expression were calculated as a fold change (fc) based on RQ values.
Statistical analysis
Allelic odds ratios (OR) and p-values, based on chi-squared (χ2) tests, were calculated using JMP Genomics 6.0 (JMP Genomics 6.0; SAS Institute Inc., Cary, NC). Deviation from Hardy-Weinberg equilibrium was analyzed using the exact test implemented in Haploview version 4.2 (available at: http://www.broad.mit.edu/haploview). For these statistical tests, p < 0.05 was considered significant.
Genes with significant association to IBD, CD, or UC were further analyzed. Group differences in gene expression were investigated using Kruskal-Wallis ANOVA or Mann–Whitney U test (Statistica 12; StatSoft Inc., Tulsa, OK). Gene expression levels were investigated in relation to genotype using Spearman’s rank correlation test (Statistica version 12.7; StatSoft Inc., Tulsa, OK) and also in relation inflammatory status, phenotype, and ongoing medical treatment using logistic regression (Statistica version 12.7; StatSoft Inc.). The analyses of the group differences of gene expressions as well as the logistic regressions were performed using ΔCT values. Box plots were created using Statistica version 12.7 (StatSoft Inc.). A Bonferroni adjusted p < 0.008 (based on the number of analyzed genes; n = 6) was considered significant for statistical analysis based on gene expression levels.
Ethical considerations
This study was approved by the ethics committee of Linköping University (dnr. M35-07, dnr. 2011/201-31). Written and informed consent were obtained from all study participants.
Results
Genetic association – case–control approach
Of the 64 SNP markers, twelve were excluded due to failed genotyping or absence of Hardy-Weinberg equilibrium (Additional file 1: Table S1). The two strongest associations were observed between IBD overall and SNP markers in MAGI2 (rs6962966; susceptibility allele A; p = 0.004) and MAGI3 (rs1343126; susceptibility allele T; p = 0.004) (Additional file 2: Table S2). These two markers were also associated with CD (p = 0.008 and p = 0.051, respectively) and UC (p = 0.045 and p = 0.011, respectively) individually, even if the association between CD and the MAGI3 SNP marker was borderline significant. Associations were also observed between IBD overall and MAGI3 (rs12119076, p = 0.022), as well as between UC and F11R (rs7546890, p = 0.043), MAGI2 (rs7803276, p = 0.031), MAGI3 (rs6689879, p = 0.043), PTEN (rs1234226, p = 0.046), and TJP1 (rs260526, p = 0.010).
Gene expression
Biopsies from different colonic segments (cecum, ascending colon, transverse colon, descending colon, sigmoid colon, and rectum) were treated as biological replicates since all genes except TJP1 were expressed at equal levels at these locations in non-IBD controls (data not shown). TJP1 showed slightly higher expression in the sigmoid colon compared to ascending colon (p = 0.006; fc = 1.16). The biopsy specimens from ileum and colon were analyzed separately.
Gene expression versus genotype
Carriage of at least one UC susceptibility allele (C) of MAGI3 rs6689879 contributed to increased MAGI3 expression level in ileal non-IBD mucosa (p = 0.002; fc = 1.7) (Table 3). No other SNP markers contributed significantly to the gene expression level of corresponding genes.
Gene expression versus inflammation
Biopsies from inflamed colonic IBD and UC mucosa expressed significant lower levels of MAGI3, PTEN, and TJP1, compared to non-inflamed colonic mucosa from the same groups of patients (Table 4). There was no overlap between the expression levels of PTEN in inflamed colonic mucosa of CD patients compared to non-inflamed CD mucosa (Fig. 3). Furthermore, biopsies from inflamed colonic IBD mucosa expressed a significantly higher level of PTPN22, compared to non-inflamed colonic IBD mucosa.
Gene expression versus phenotype
The gene expression levels of analyzed genes were not different between any of the phenotypes (CD, UC, or non-IBD) (Additional file 3: Table S3).
Gene expression versus medication
Expression of investigated genes in non-inflamed and inflamed IBD biopsy specimens was not affected by ongoing medical treatment (thiopurines, aminosalicylates, or glucocorticosteroids) (Additional file 4: Table S4). Too few individuals underwent anti-TNF-\( \alpha \)-antibody treatment for meaningful statistical analysis.
Discussion
IBD has been associated with an increased paracellular permeability of the intestinal epithelium and a dysfunctional barrier is considered important in the pathogenesis of IBD [6, 7]. The TJ structure is critical for the paracellular barrier properties [9]. Although a few IBD susceptibility markers have been linked to barrier related genes, e.g. CLDN2 and MAGI2 [11, 12, 20], the extent to which a dysfunctional barrier is a consequence of genetic factors remains unclear.
The large numbers of susceptibility loci identified in relation to IBD [3,4,5] only explain 10.9% and 7.7% of the heritability of CD and UC, respectively [3]. Regional heterogeneity of allele frequencies among different sub-populations has been observed [4, 5, 21], and indicates the importance of identifying associations in homogenous populations to increase study power [21] and to explain the genetic variance in IBD. To further clarify the role of the paracellular barrier in the etiology of IBD, a Swedish population of IBD patients and non-IBD controls was genotyped for several TJ related genes and analyzed for expression in the intestinal mucosa.
One strong association was observed between the T-allele of the MAGI3 SNP marker rs1343126 and IBD, CD, and UC. Further, the C-allele of another MAGI3 SNP marker (rs6689879) was found to be significantly associated with UC. Presence of this marker also resulted in an increased MAGI3 expression in ileal non-IBD mucosa, which might contribute to the restriction of inflammation in UC to the colon. To the best of our knowledge we have for the first time demonstrated an association between MAGI3 and IBD. However, Jostins et al. have previously demonstrated association between this genetic region and CD, but primarily highlighted PTPN22 [5].
In our study, carriage of at least one IBD associated allele at rs1343126 was nominally associated to a decreased colonic expression of PTPN22 (located in the same genetic region as MAGI3) in the non-IBD subgroup. The corresponding relationship was not identified in inflamed or non-inflamed mucosa of IBD patients. In fact, all relationships between gene expression levels and the susceptibility alleles were observed in mucosal biopsies from non-inflamed, non-IBD controls. Even though the expression of several genes were affected in inflamed mucosa, it is unlikely that genotype – gene expression relationships were concealed in the IBD patients by clinical or subclinical inflammation in the non-inflamed biopsies, since these biopsies were obtained from IBD mucosa that were assessed as histopathologically normal. It is, however, possible that the investigated genes were affected by other factors (e.g. microRNA) that persist in non-inflamed IBD-mucosa [22,23,24].
Jostins et al. previously identified the C-allele of rs6679677 (MAGI3-PTPN22) as a susceptibility allele of CD, but a protective allele with respect to UC [5], which was not confirmed by our study. Nevertheless, it is possible that our study missed a possible association due to a comparatively small sample size.
Significant associations were observed between an SNP marker in MAGI2 (rs6962966) and IBD, CD, as well as UC. Wapenaar et al. [11] previously described an association between this marker and UC. Their study revealed, however, an association to the opposite allele (G) compared to our study (A). Presently we have no explanation for this discrepancy.
In our association study we applied a significance level of p < 0.05, and the findings were further followed up by analyzing for a genotype-gene expression correlation in a second independent cohort. However, even if we apply a more stringent p-value, by making a Bonferroni adjustment based on the number of analyzed genes (n = 8; p < 0.006), both MAGI3 (rs1343126) and MAGI2 (rs6962966) would still remain significantly associated to IBD.
One limitation of our study was that MAGI1 and MAGI3 were only investigated for SNP markers of exons and exon-intron-boundaries, due to the large size of these genes, and that the selection of SNP markers of MAGI2, MAGI3-PTPN22, PARD3, and TJP1 were based on reports in the literature.
MAGI3 was expressed to a lower level in inflamed, compared to non-inflamed, colonic mucosa from IBD and UC patients. MAGI3 is involved in suppression of the PI3K/Akt pathway due to interaction with PTEN [25], and also in the Wnt/β-catenin pathway [26, 27]. T cells with a high β-catenin level induce T cell activation and intestinal inflammation [28]. Thus, regulation of these pathways might constitute a mechanism by which decreased MAGI3 expression promotes intestinal inflammation. Furthermore, the PTPN22 gene expression was shown to be increased in inflamed colonic IBD mucosa, compared to non-inflamed mucosa, which corroborates previous results reported by Chen et al. [29]. The PTPN22 gene is expressed at a higher level in immune cells compared to non-immune cells [30], and further PTPN22 negatively regulates T cell receptor signaling [31]. In our study gene expression was determined using biopsies, and therefore we are unable to determine the exact cellular origin of the increased expression in inflamed mucosa. It may, however, be due to increased quantities of PTPN22-expressing immune cells during inflammation.
Decreased PTEN expression was observed in inflamed colonic mucosa of IBD, CD, and UC patients, compared to non-inflamed mucosa. Furthermore, there was no overlap between the PTEN expression levels in inflamed colonic mucosa of CD patients, compared to non-inflamed CD mucosa, suggesting PTEN as a marker for inflammatory response in colonic CD mucosa. PTEN is described to play a role in the regulation of intestinal permeability [32, 33], and is also defined as a tumor suppressor molecule inhibiting inflammatory response, cell migration, and proliferation via PI3K/Akt pathway [34, 35]. An increased level of microRNA-21 has been described in colonic biopsies from UC patients, compared to normal colonic mucosa, and expression of microRNA-21 in the Caco-2 cell line, a colonic epithelial cell model, resulted in an increased permeability [36]. The mechanism of microRNA-21 has been investigated in Caco-2 cells, and may involve an effect on paracellular permeability via the PTEN level and Akt phosphorylation [33]. Moreover, activation of PI3K/Akt pathway has been demonstrated to inhibit the differentiation of T cells towards regulatory T cells [37] and previously a decreased PTEN expression level was observed in intestinal mucosal lymphocytes in CD patients, compared to controls [38].
The decreased TJP1 expression level observed in inflamed IBD and UC mucosa, compared to non-inflamed mucosa, is consistent with a loss of ZO-1 protein (encoded by TJP1) in dextran sulfate sodium induced colitis in mice [39]. Poritz et al. further observed a correlation between loss of ZO-1 and increased paracellular permeability [39].
Gene expression was not affected by ongoing medical treatment (thiopurines, aminosalicylates, or glucocorticosteroids), irrespective of the presence of inflammation. Nevertheless, other studies have shown that infliximab, an anti-TNF-antibody, can impact the expression of genes associated with intestinal permeability [40] and mucosal cytokine expression [41]. Among our patients, too few were treated with infliximab to allow meaningful analysis with this drug.
Conclusions
In conclusion, our findings support a genetic basis for modulation of the intestinal epithelial barrier in IBD, and point to a complex regulation of TJ gene expression through a secondary role of inflammation. By focusing on a Swedish population and a network of interacting TJ components we have identified MAGI3 as a new candidate gene for IBD.
Abbreviations
- CD:
-
Crohn’s disease
- CT :
-
Threshold cycle
- Fc:
-
Fold change
- IBD:
-
Inflammatory bowel disease
- OR:
-
Odds ratio
- RQ:
-
Relative quantification
- SNP:
-
Single nucleotide polymorphism
- TJ:
-
Tight junctions
- UC:
-
Ulcerative colitis
References
Peloquin JM, Nguyen DD. The microbiota and inflammatory bowel disease: insights from animal models. Anaerobe. 2013;24:102–6.
Loddo I, Romano C. Inflammatory Bowel Disease: Genetics, Epigenetics, and Pathogenesis. Front Immunol. 2015;6:551.
Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48(5):150–8.
Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47(9):979–86.
Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–24.
John LJ, Fromm M, Schulzke JD. Epithelial barriers in intestinal inflammation. Antioxid Redox Signal. 2011;15(5):1255–70.
Lee SH. Intestinal permeability regulation by tight junction: implication on inflammatory bowel diseases. Intestinal res. 2015;13(1):11–8.
Hollander D, Vadheim CM, Brettholz E, Petersen GM, Delahunty T, Rotter JI. Increased intestinal permeability in patients with Crohn’s disease and their relatives. A possible etiologic factor. Ann Intern Med. 1986;105(6):883–5.
Vetrano S, Danese S. The role of JAM-A in inflammatory bowel disease: unrevealing the ties that bind. Ann N Y Acad Sci. 2009;1165:308–13.
Buhner S, Buning C, Genschel J, Kling K, Herrmann D, Dignass A, et al. Genetic basis for increased intestinal permeability in families with Crohn's disease: role of CARD15 3020insC mutation? Gut. 2006;55(3):342–7.
Wapenaar MC, Monsuur AJ, van Bodegraven AA, Weersma RK, Bevova MR, Linskens RK, et al. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. 2008;57(4):463–7.
McGovern DP, Taylor KD, Landers C, Derkowski C, Dutridge D, Dubinsky M, et al. MAGI2 genetic variation and inflammatory bowel disease. Inflamm Bowel Dis. 2009;15(1):75–83.
KEGG PATHWAY Database. Available at: http://www.genome.jp/kegg/pathway.html (Accessed 19 November 2015).
STRING. Available at: http://string-db.org/. Accessed 16 Aug 2016.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–452.
Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43(11):1066–73.
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9.
Söderman J, Berglind L, Almer S. Gene Expression-Genotype Analysis Implicates GSDMA, GSDMB, and LRRC3C as Contributors to Inflammatory Bowel Disease Susceptibility. Biomed Res Int. 2015;2015:834805.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8.
Soderman J, Noren E, Christiansson M, Bragde H, Thiebaut R, Hugot JP, et al. Analysis of single nucleotide polymorphisms in the region of CLDN2-MORC4 in relation to inflammatory bowel disease. World J gastroenterol : WJG. 2013;19(30):4935–43.
Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17(R2):R143–150.
Fasseu M, Treton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS one. 2010;5(10):e13160. doi:10.1371/journal.pone.0013160.
Planell N, Lozano JJ, Mora-Buch R, Masamunt MC, Jimeno M, Ordas I, et al. Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut. 2013;62(7):967–76.
Peloquin J, Goel G, Huang H, Haritunians T, Sartor R, Daly M, et al. O-002 Genes in IBD-Associated Risk Loci Demonstrate Genotype-, Tissue-, and Inflammation-Specific Patterns of Expression in Terminal Ileum and Colon Mucosal Tissue. Inflamm Bowel Dis. 2016;22 Suppl 1:S1.
Wu Y, Dowbenko D, Spencer S, Laura R, Lee J, Gu Q, et al. Interaction of the tumor suppressor PTEN/MMAC with a PDZ domain of MAGI3, a novel membrane-associated guanylate kinase. J Biol Chem. 2000;275(28):21477–85.
Ma Q, Zhang Y, Meng R, Xie KM, Xiong Y, Lin S, et al. MAGI3 Suppresses Glioma Cell Proliferation via Upregulation of PTEN Expression. Biomed environ sci : BES. 2015;28(7):502–9.
Ma Q, Yang Y, Feng D, Zheng S, Meng R, Fa P, et al. MAGI3 negatively regulates Wnt/beta-catenin signaling and suppresses malignant phenotypes of glioma cells. Oncotarget. 2015;6(34):35851–65.
Keerthivasan S, Aghajani K, Dose M, Molinero L, Khan MW, Venkateswaran V, et al. beta-Catenin promotes colitis and colon cancer through imprinting of proinflammatory properties in T cells. Sci Transl Med. 2014;6(225):225–8.
Chen Z, Zhang H, Xia B, Wang P, Jiang T, Song M, et al. Association of PTPN22 gene (rs2488457) polymorphism with ulcerative colitis and high levels of PTPN22 mRNA in ulcerative colitis. Int J Color Dis. 2013;28(10):1351–8.
Arimura Y, Yagi J. Comprehensive expression profiles of genes for protein tyrosine phosphatases in immune cells. Sci Signal. 2010;3(137):rs1.
Sharp RC, Abdulrahim M, Naser ES, Naser SA. Genetic Variations of PTPN2 and PTPN22: Role in the Pathogenesis of Type 1 Diabetes and Crohn’s Disease. Front Cell Infect Microbiol. 2015;5:95.
Langlois MJ, Bergeron S, Bernatchez G, Boudreau F, Saucier C, Perreault N, et al. The PTEN phosphatase controls intestinal epithelial cell polarity and barrier function: role in colorectal cancer progression. PLoS One. 2010;5(12):e15742.
Zhang L, Shen J, Cheng J, Fan X. MicroRNA-21 regulates intestinal epithelial tight junction permeability. Cell Biochem Funct. 2015;33(4):235–40.
Hu Y, Li Z, Guo L, Wang L, Zhang L, Cai X, et al. MAGI-2 Inhibits cell migration and proliferation via PTEN in human hepatocarcinoma cells. Arch Biochem Biophys. 2007;467(1):1–9.
Tokuhira N, Kitagishi Y, Suzuki M, Minami A, Nakanishi A, Ono Y, et al. PI3K/AKT/PTEN pathway as a target for Crohn’s disease therapy (Review). Int J Mol Med. 2015;35(1):10–6.
Yang Y, Ma Y, Shi C, Chen H, Zhang H, Chen N, et al. Overexpression of miR-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the Rho GTPase RhoB. Biochem Biophys Res Commun. 2013;434(4):746–52.
Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A. 2008;105(22):7797–802.
Long SH, He Y, Chen MH, Cao K, Chen YJ, Chen BL, et al. Activation of PI3K/Akt/mTOR signaling pathway triggered by PTEN downregulation in the pathogenesis of Crohn’s disease. J Dig Dis. 2013;14(12):662–9.
Poritz LS, Garver KI, Green C, Fitzpatrick L, Ruggiero F, Koltun WA. Loss of the tight junction protein ZO-1 in dextran sulfate sodium induced colitis. J Surg Res. 2007;140(1):12–9.
Toedter G, Li K, Sague S, Ma K, Marano C, Macoritto M, et al. Genes associated with intestinal permeability in ulcerative colitis: changes in expression following infliximab therapy. Inflamm Bowel Dis. 2012;18(8):1399–410.
Arijs I, De Hertogh G, Machiels K, Van Steen K, Lemaire K, Schraenen A, et al. Mucosal gene expression of cell adhesion molecules, chemokines, and chemokine receptors in patients with inflammatory bowel disease before and after infliximab treatment. Am J Gastroenterol. 2011;106(4):748–61.
Acknowledgments
We would like to thank laboratory technician Lena Svensson (Department of Clinical and Experimental Medicine, Linköping University, Sweden) for excellent technical assistance and support.
Funding
This work was supported by FORSS, the Medical Research Council of South-Eastern Sweden [grant No 236541-2012 and 235131-2012], Futurum - the Academy for Healthcare, Region Jönköping County [grant No FUTURUM-338631], Bengt Ihre-Fonden [grant No 2012-SLS 254491], and Karolinska Institutets Forskningsfonder [grant No 2014fobi42063]. The funding bodies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
All authors conceived of the study; all authors designed the study; EN contributed to all laboratory work; EN contributed to data analysis and drafted the manuscript; SA provided blood samples and patient data; EN performed the statistical analysis; all authors contributed to the context of the manuscript; all authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by the ethics committee of Linköping University (dnr. M35-07, dnr. 2011/201-31). Written and informed consent were obtained from all study participants.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
Single nucleotide polymorphism markers included in the study. (XLSX 16 kb)
Additional file 2: Table S2.
Inflammatory bowel disease-phenotypes were investigated by performing case–control approach in genetic association studies. (XLSX 24 kb)
Additional file 3: Table S3.
For genes with a significant genetic association to IBD, CD, or UC, gene expression was analyzed in relation to phenotype, using logistic regression. (DOCX 16 kb)
Additional file 4: Table S4.
For genes with a significant genetic association to IBD, CD, or UC, gene expression was analyzed in relation to medical treatment, using logistic regression. (DOCX 17 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Norén, E., Almer, S. & Söderman, J. Genetic variation and expression levels of tight junction genes identifies association between MAGI3 and inflammatory bowel disease. BMC Gastroenterol 17, 68 (2017). https://doi.org/10.1186/s12876-017-0620-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12876-017-0620-y