Abstract
Background
The development of CRISPR/Cas9 technology has facilitated targeted mutagenesis in an efficient and precise way. Previously, RNAi silencing of the susceptibility (S) gene PowderyMildewResistance 4 (PMR4) in tomato has been shown to enhance resistance against the powdery mildew pathogen Oidium neolycopersici (On).
Results
To study whether full knock-out of the tomato PMR4 gene would result in a higher level of resistance than in the RNAi-silenced transgenic plants we generated tomato PMR4 CRISPR mutants. We used a CRISPR/Cas9 construct containing four single-guide RNAs (sgRNAs) targeting the tomato PMR4 gene to increase the possibility of large deletions in the mutants. After PCR-based selection and sequencing of transformants, we identified five different mutation events, including deletions from 4 to 900-bp, a 1-bp insertion and a 892-bp inversion. These mutants all showed reduced susceptibility to On based on visual scoring of disease symptoms and quantification of relative fungal biomass. Histological observations revealed a significantly higher occurrence of hypersensitive response-like cell death at sites of fungal infection in the pmr4 mutants compared to wild-type plants. Both haustorial formation and hyphal growth were diminished but not completely inhibited in the mutants.
Conclusion
CRISPR/Cas-9 targeted mutagenesis of the tomato PMR4 gene resulted in mutants with reduced but not complete loss of susceptibility to the PM pathogen On. Our study demonstrates the efficiency and versatility of the CRISPR/Cas9 system as a powerful tool to study and characterize S-genes by generating different types of mutations.
Similar content being viewed by others
Background
Powdery mildew (PM) in tomato, caused by the obligate biotrophic fungus Oidium neolycopersici (On), is a world-wide disease that threatens the production of greenhouse- and field-grown tomatoes [1, 2]. Over the last few decades, research focused on breeding for resistance against PM in tomato has resulted in the identification of five dominant resistance (R) genes (Ol-genes) from wild tomato species [3]. These genes were introgressed into the susceptible tomato cultivar Moneymaker and near-isogenic lines (NILs) were made [4]. Histological studies of these NILs after powdery mildew infection has allowed the identification of two different types of host responses associated with resistance: unicellular hypersensitive response (HR) in Ol-4 and Ol-6, leading to complete resistance against PM; and slow multicellular HR in Ol-1, Ol-3 and Ol-5, leading to incomplete resistance [4,5,6].
In addition to these dominantly-inherited resistance genes a recessive gene named ol-2 was identified [7]. This is a loss-of-function mutant allele of the powdery mildew susceptibility (S) gene, MILDEW RESISTANCE LOCUS O (MLO) [7]. In homozygous state this ol-2 allele confers broad-spectrum resistance to different PM species by inducing papilla formation and callose deposition, which block fungal development at penetration stage [6]. The loss-of-function allele of MLO is one of the best studied examples of recessively-inherited resistance against PMs [8]. In recent years, many other examples of resistance conferred by the impairment of S genes have been described in different pathosystems [9]. The use of such S genes in plant breeding, due to their broad-spectrum and potentially durable resistance characteristics, represent a promising alternative to the introgression of R genes that has been driving traditional resistance breeding [9,10,11].
In Arabidopsis, several mutants showing resistance against the adapted PM pathogen Golovinomyces cichoracearum have been identified after screening an EMS population [12]. These mutants were named powdery mildew resistant 1 (pmr1) to pmr4. The pmr4 mutant showed resistance not only against G. cichoracearum, but also against G. orontii and downy mildew Hyaloperonospora arabidopsidis. PMR4 was shown to be the callose synthase gene At4g03550, also known as GLUCAN SYNTHASE-LIKE 5 (GSL5) or CALLOSE SYNTHASE 12 (CalS12) [13, 14]. The GSL/CalS gene family comprises 12 genes in Arabidopsis [15,16,17]. The callose synthase encoded by PMR4 is responsible for the production of callose in response to biotic and abiotic stresses [14]. Both the pmr4 and a gsl5 mutant (homozygous for a T-DNA insertion in the PMR4 gene) showed almost complete lack of callose in infected leaves [12, 13]. Histological analyses proved that these mutants form papillae after PM infection, even in the absence of callose [13, 14].
It would seem strange that mutant plants lacking callose in the papillae formed at attempted penetration sites show PM resistance, especially because Arabidopsis plants overexpressing PMR4 show complete penetration resistance against G. cichoracearum [18, 19]. This latter resistance is based on increased callose deposition at infection sites, which acts as a physical barrier against PM-secreted cell wall hydrolases [19].
However, the pmr4 mutant develops lesions reminiscent of hypersensitive cell death after PM infection [14]. This cell death likely results from activation of the salicylic acid (SA) signal transduction pathway as genes in this pathway were upregulated in the pmr4 mutant compared to the wild type control, and blocking the SA pathway in the pmr4 mutant (using double mutants) was enough to restore full susceptibility to PM [14]. Probably, PMR4 is not only involved in callose synthesis at specific sites after attempted fungal penetration, but also negatively controls the SA-associated defense pathway [20]. In plants overexpressing PMR4 the SA pathway is not induced [18]. Thus, different mechanisms are involved in resistance by overexpressing or knocking-out the PMR4 gene.
To investigate whether the PMR4 function is conserved in other plants species than Arabidopsis, the closest tomato ortholog of PMR4 (Solyc07g053980; SlPMR4) was silenced by RNAi [21]. Transgenic plants in which the gene was well-silenced showed enhanced resistance against the tomato PM pathogen (On) and a slight reduction in plant size when compared to the non-silenced controls [21]. These RNAi plants still showed a low level of expression of the SlPMR4 gene. Knock-down-based methods for characterization of S genes, such as RNAi or virus-induced gene silencing (VIGS), usually result in residual expression of the targeted genes, which typically causes partial phenotypes complicating functional analysis of the genes [22]. Mutations resulting in full knock-out of gene function sometimes produce a stronger phenotype than knock-down transgenic plants obtained by RNAi silencing [23, 24] . We set out to produce tomato SlPMR4 mutants to investigate whether complete resistance against On could be obtained in this way, compared with the substantial but not complete resistance in the RNAi transgenic plants [21] . For this, targeted mutagenesis is the preferred method. The generation of precise, stable and heritable knock-out alleles of S genes is now possible with the development of the clustered regularly interspaced short palindromic repeats (CRISPR) technology. This technology has already been used to generate PM-resistant slmlo1 tomato mutants [25], and PM-resistant wheat by the simultaneous modification of the three EDR1 homologues [26]. In addition, host resistance in other pathosystems has been achieved using CRISPR-induced targeted mutation [27, 28]. In this study, we report the successful generation and characterization of five different mutation events via CRISPR/Cas9 in the tomato ortholog of the PMR4 gene. Further, we show that PM resistance in these pmr4 mutants is associated with cell death upon PM infection.
Results
CRISPR/Cas-9-targeted mutagenesis of SlPMR4
To produce mutants of the tomato PMR4 ortholog (Solyc07g053980) [21] a single CRISPR/Cas-9 construct containing four sgRNAs (sgRNA6, sgRNA8, sgRNA1 and sgRNA7; Supplementary Table 1) was made. We used four sgRNAs in order to increase the chances of obtaining mutants with large deletions [29, 30] . The position of the four sgRNAs in the SlPMR4 genomic sequence and in the predicted SlPMR4 protein can be seen in Fig. 1. PMR4 contains two known protein domains according to NCBI Conserved Domains (https://www.ncbi.nlm.nih.gov/cdd/): FKS1dom1 [32] and the Glucan-synthase domain [33]. One of the four sgRNAs (sgRNA6) targets the FKS1dom1 domain in the N-terminal region. The three remaining sgRNAs target the intracellular part of the Glucan-synthase domain. The SlPMR4 CRISPR/Cas9 construct was used to transform the susceptible tomato cultivar Moneymaker (MM).
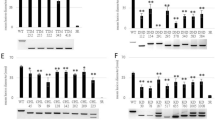
Position of target sites of the sgRNAs in SlPMR4. a. Representation of genomic sequence of SlPMR4 showing the position of four single guide RNAs (sgRNAs) that were designed to produce knock-out mutants of SlPMR4. PCR primers Fw519 + Rv1925 and Fw2969 + Rv4230 were used to identify mutants. b. PROTTER [31] representation of the SlPMR4 protein. sgRNA6 (green) is located in the N-terminal FKS1dom1 domain of the protein; sgRNA8 (yellow), sgRNA1 (orange) and sgRNA7 (red) target sites in the intracellular part of the Glucan-synthase domain
In total, 37 primary transformants (T1) were obtained and analyzed. Individual T1 plants containing mutations within the SlPMR4 gene were selected via PCR amplification. Two different primer combinations were used: primers Fw519 + Rv1925 flanking sgRNA6 and primers Fw2969 + Rv4230 flanking sgRNA8, sgRNA1 and sgRNA7 (Fig. 1). When a large deletion has occurred smaller PCR products than the expected ones from wild-type (WT) controls will be visible on an agarose gel. Additionally, the sgRNA6 target site plus PAM contains the recognition sequence for restriction enzyme XcmI (CCANNNNN↓NNNNTGG). When a small indel has occurred within this target site the PCR product will not be digested by XcmI.
The PCR with primers Fw519 + Rv1925 flanking sgRNA6 (Fig. 1) did not yield PCR products obviously smaller than the 1407-bp WT PCR product in any of the primary transformants, indicating no large deletions had occurred in this region. In contrast, PCR with primers Fw2969 + Rv4230 flanking the three other sgRNAs resulted in the selection of 17 T1 transformants with large deletions compared to the 1262-bp PCR product of the WT allele. These were subsequently selfed to produce T2 progeny. T2 progenies were analyzed by PCR and individual T2 plants homozygous for the mutation were selected to produce T3 progenies to facilitate further characterization of the mutations (results shown in the next section).
To analyze whether any of the selected T2 or T3 mutants contained a small indel at the sgRNA6 target site PCR products obtained with primers Fw519 + Rv1925 were digested with XcmI. For all tested plants digestion was complete, suggesting that no mutation had occurred at the sgRNA6 target site. To be sure none of the mutants contained a mutation in the sgRNA6 region the PCR products were sequenced. No differences with the WT allele were observed. These results indicate that sgRNA6 in the FKS1dom1 domain was not effective in producing mutations, whereas one or more of the three sgRNAs in the Glucan-synthase domain were successful.
Characterization of CRISPR/Cas-9-mediated mutants in SlPMR4
We characterized the mutation events in T2 and T3 progenies derived from the original transgenic T1 plants (Table 1). From segregating T2 families plants homozygous for potential mutant alleles were selected that showed size differences in the PCR amplified products (using primers Fw2969 + Rv4230) when compared to the WT allele. In addition, homozygous T3 lines were obtained and the presence of mutant alleles was verified by repeating the PCR amplification (Fig. 2).
PCR amplification of the CRISPR tomato mutants. Selection of the mutants was done by amplifying the region containing sgRNAs 8, 1 and 7 using flanking primers Fw2969 and Rv4230 (Fig. 1). A 1262-bp wild-type allele was amplified in Moneymaker (MM) and transformant TV161196 (non-mutant). Smaller fragments than the wild-type allele indicate deletions between sgRNAs. Different mutation events are indicated and sequence details of events 1 to 5 are given in Fig. 3
By sequencing of PCR fragments we identified five different mutation events (“events”) in the mutant lines (Fig. 3, Table 1 and Supplementary Document 1). Event 1 contains a 900-bp in-frame deletion between sgRNA8 and sgRNA7. Event 2 shows deletions of 5 bp at sgRNA8 and 277 bp at sgRNA7. Event 3 carries a 895-bp deletion between sgRNA8 and sgRNA7. Event 4 has a 902-bp deletion between sgRNA8 and sgRNA7 and the insertion of a T at the site of the mutation. Event 5 is a special case that has a 4-bp deletion and a 892-bp inversion next to the deletion between sgRNA8 and sgRNA7. With the exception of event 1, all the mutation events are predicted to generate premature translation termination codons (PTTCs) in the transcript (Supplementary figure1). The predicted protein of event 1 is lacking 300 amino acids in the Glucan-synthase domain.
Schematic representation of the mutation events in SlPMR4 mutants. Two exons (E1 and E2) of the SlPMR4 gene are shown. The positions of the four sgRNAs are shown in red. The mutation for each event is represented at the genomic level. Deletions ranging from 4-bp to 902-bp were observed. Additionally, a 892-bp inversion is present in the event 5 mutants. * indicates the presence of premature translation termination codons (PTTCs) in the predicted protein
Phenotypically, the CRISPR/Cas9 pmr4 mutants were similar to non-transformed MM. However, they displayed a slight reduction in size when compared to MM and WT allele-carrying transformants. An exception to this is line TV171370 that showed segregation of plants with a dwarf phenotype (three out of eight T3 progeny). To investigate the cause of this dwarf phenotype a PCR was performed to check for the presence of the Cas9 sequence and possible association with T-DNA integration site. All eight plants carried the Cas9 gene; therefore no association was found with T-DNA integration. Next, the sgRNA6 target site was investigated by sequencing of the Fw519 + Rv1925 PCR product. No additional mutation(s) were observed in the dwarf plants.
One of the criteria for selection of the sgRNAs was a minimal chance of off-targets (unintended mutations elsewhere in the genome instead of SlPMR4). When using the Cas-OFFinder program [34] with a threshold of 3 or less mismatches between sgRNA and (off-) target sequence no putative off-targets were predicted for sgRNA 6, one for sgRNA8 (in an intergenic region), one for sgRNA1 (in an intron of gene Solyc09g009800) and three for sgRNA7 (Supplementary Table 2). These off-targets all contain 3 mismatches, most of which are present in the seed of the sgRNA. From the three off-targets of sgRNA7 two are in intergenic regions and one in the coding sequence of gene Solyc02g078230. This gene encodes a callose synthase-like gene designated SlPMR4-h2 [21]. Because of this we investigated whether any mutation had occurred in the SlPMR4-h2 gene in our selected SlPMR4(−h1) mutants. For this, a PCR was performed using primers PMR4_h2_Fw1 and PMR4_h2_Rv1 yielding a 765-bp PCR product containing the putative off-target site of sgRNA7. In total, PCR products of 36 individual T2 and T3 mutant plants and 6 control plants (MM and WT-allele carrying transgenic plants) were sequenced. No sequence differences were found between control and mutant (including the three dwarf) plants, indicating no off-target mutation had occurred in the SlPMR4-h2 gene.
Resistance to powdery mildew in slpmr4 mutants
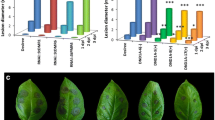
A previous study with RNAi lines showed that the knock-down of SlPMR4 enhances resistance against powdery mildew [21]. To evaluate whether our slpmr4 mutant lines showed increased or full resistance against PM, we inoculated them with On to assess the disease index (Fig. 4a and b). Additionally, we quantified the disease severity by measuring the relative On biomass in the mutants to confirm the phenotypic observations (Fig. 4c). Two unsuccessful transgenic lines (TV161196 and TV161209) carrying the WT allele were used as controls. No significant differences in the disease index or the relative fungal biomass were observed among the mutants. However, all the mutants displayed reduced susceptibility compared to the controls as indicated by a lower disease index and significantly lower fungal biomass (Fig. 4, Supplementary figure2).
Phenotypic response of the pmr4 mutants to infection with Oidium neolycopersici. a. Powdery mildew symptoms observed on the leaves of wild-type plants and mutants (one genotype is given from each of the five mutation events). Photos were taken at 21 days post inoculation (dpi). b. Average disease index score of the mutant lines at 10 and 12 dpi. c. Relative fungal biomass quantification on at least three individual plants of the mutant lines. This is calculated as the ratio of fungal ITS gene amplification in comparison with tomato EF1a and normalized with the values of the wild-type genotype TV161196. Significant differences were observed between the different mutation events and the control lines (Tukey HSD test, α = 0.05)
Histological analysis of the infection sites of Arabidopsis pmr4 mutants revealed the presence of hypersensitive response (HR)-like cell death [14]. To investigate whether this phenomenon could also be observed in our slpmr4 mutants, we performed histological studies using heavily infected leaf samples of 21 plants representing all five mutation events in addition to wild-type MM plants. The samples were taken at 44 h post infection (hpi) and stained with trypan blue. Fungal structures and plant cell death were quantified in both mutant and wild-type plants (Table 2). No papillae were observed in any of the samples. HR-like cell death was visible at a much higher percentage of the infection sites in all pmr4 mutants compared to the wild-type allele-carrying MM (Table 2 and Fig. 5). Simultaneously, the percentage of primary haustoria and the number of hyphae per infection unit was decreased in the mutants compared to MM. These results corroborate the finding that tomato pmr4 mutants show reduced susceptibility to On, but not complete resistance.
Microscopic observations on powdery mildew infection at 44 h post inoculation (hpi). a. In the wild-type allele-carrying plant (TV161209) a normal development of the spores occurs; appressorium and hyphae are developed. b,c. In the mutant plant (TV171010; event 4) cell death is observed in epidermal cells invaded by the fungus. c shows a deeper focal plane involving the same infection unit as in b; a haustorium is present in the cell showing cell death. d. Both cell death and hyphal growth in mutant plant TV171010. e. Comparison of number of hyphae per infection unit between wild type and pmr4 mutant plants
In Arabidopsis it was shown that PM resistance in the pmr4 mutant is associated with an activation of the SA signal transduction pathway [14]. To analyze whether the SA pathway is also activated in our slpmr4 mutants expression level of the tomato PR1 gene 44 h after PM infection was determined by qPCR. Infected leaf samples were taken from individual homozygous mutant T2 plants representing all five mutation events and control plants (MM and unsuccessful transgenic lines TV161196 and TV161209). Disease index scores of this experiment are shown in Fig. 6a and relative PR1 gene expression in Fig. 6b. Plants from all five mutation events show reduced susceptibility (but not complete resistance) to PM compared to the control lines (Fig. 6a). All mutant plants showed a significant increase in PR1 gene expression at 44 h post infection (hpi) compared with the non-mutant controls (Fig. 6b).
Discussion
Effectiveness of multiple guide RNAs to obtain knock-out mutants
The use of loss-of-function alleles of S-genes in plant breeding is a promising alternative to the traditional R-gene-based introgression breeding because of its durable and broad-spectrum characteristics [10]. In this study, we successfully produced CRISPR/Cas-9-mediated knock-outs of the susceptibility gene PMR4 in tomato against the PM pathogen On. Our results showed that the use of four sgRNAs for CRISPR-induced mutation and selection using PCR amplification to screen for visible (large) deletions was efficient to obtain the five described mutation events. Analysis of the different target sites indicated a difference in effectiveness of the four sgRNAs although all four sgRNAs were selected based on the same stringent criteria such as GC content, secondary structure and base pairing scores (Supplementary Table 1). No mutations were found close to the sgRNA6 target site, thus it was not efficient in guiding the Cas9 protein to induce double-stranded breaks. However, all five described mutation events seem to be the result of double-stranded breaks at target sites of both sgRNA8 and sgRNA7, as deletions were found at or between these positions. Therefore, sgRNA8 and sgRNA7 seem to be highly effective. We cannot judge whether sgRNA1 was effective, as the target site of this sgRNA is positioned between those of sgRNA8 and sgRNA7, and was deleted in three of the five mutation events (Fig. 3). As we focused on the selection of mutants with large deletions in the SlPMR4 gene we have not checked whether any additional mutants with small indels (insertion or deletions of a few nucleotides) at the targets sites of sgRNA1, 7 or 8 were present among the original primary transformants. Such an analysis would allow a better comparison of the effectiveness of sgRNAs1, 7 and 8.
One of the five mutation events we characterized contained a 892-bp inversion, showing that chromosome re-arrangement can occur by using CRISPR/Cas9 technology. In agreement with our results, genome editing via the CRISPR/Cas9 system has recently been reported to produce inversions between sgRNAs [35, 36]. Induction of inversions are of particular interest in plant breeding due to its potential to allow the fixing or breaking of linkages [36].
Putative pleiotropic effects of SlPMR4 mutation
Similar to the observation on transgenic plants in which SlPMR4 (Solyc07g053980) was silenced by RNAi [21], a slight reduction in plant size was observed in the CRISPR/Cas9 slpmr4 mutants. This may be due to an elevated SA level in the mutants. Constitutively high levels of SA can result in growth impairment in Arabidopsis [37, 38]. Although we did not measure SA levels in our mutant lines, expression of the PR1 gene (as an indicator of SA levels) was increased in PM-infected leaves of the slpmr4 mutants compared to controls containing WT alleles of SlPMR4.
Three out of eight individuals of T2 family TV171370 showed a dwarf phenotype. We tried to identify the cause for this dwarf phenotype. None of the plants showed additional mutations at the sgRNA6 target site of SlPMR4 or at the possible off-target site in Solyc02g078230 (SlPMR4-h2). Furthermore, all eight plants were positive for the Cas9 gene. Therefore, no obvious cause for the occurrence of dwarf plants was identified. However, it is still possible that the site of T-DNA integration plays a role. When the T-DNA of the CRISPR/Cas9 construct has integrated within a gene influencing plant growth, dwarf plants are obtained only when the T-DNA insertion is present in homozygous state. Another possibility is that multiple integrations of the T-DNA had occurred in the parental T1 plant of TV171370, one of which would be in a crucial gene for plant development.
As dwarf plants were only found in one T2 family and not in any of the others this phenotype does not seem to be caused by mutation of the SlPMR4 gene. The possible pleiotropic effects in loss-of-susceptibility lines have been discussed as a setback to the deployment of S-genes in plant breeding [9]. Clearly, further phenotypic analyses in standard greenhouse conditions and yield estimation should determine whether the gain in resistance of pmr4 tomato plants also carries a fitness cost in the plants.
Resistance to PM in knock-out SlPMR4 plants
We aimed to investigate whether the full knock-out of SlPMR4 would result in a higher level of resistance than RNAi-silenced transgenic plants [21] . In this study we have shown that our slpmr4 mutants in the susceptible cultivar Moneymaker background displayed enhanced resistance to tomato PM compared to control plants containing WT alleles of SlPMR4, but not complete resistance. We evaluated the plants for disease index at 10–17 dpi (Figs. 4 and 6) and determined fungal biomass at 21 dpi. It could be argued that susceptibility in the mutants is not reduced but delayed. However, we kept the inoculated pmr4 mutants till 30–35 dpi and still the plants showed less infection than the WT control plants. Therefore, the pmr4 mutants show reduced susceptibility to PM.
In agreement with previous observations in Arabidopsis pmr4 mutants [14] we observed a higher occurrence of HR-like cell death at infection sites in tomato CRISPR pmr4 mutants compared to wild type plants. This increased HR likely resulted from activation of the SA signaling pathway, as PR1 gene expression was significantly increased in the pmr4 mutants compared to the controls. Still, this cell death did not completely block fungal growth in the tomato pmr4 mutants. In barley, RNAi was used to downregulate gene HvGsl6, which is the closest ortholog of AtGSL5 (PMR4) [39]. As expected, this resulted in a lower accumulation of papillary and wound callose. However, contrary to what has been found in Arabidopsis and tomato, silencing of HvGsl6 led to a higher susceptibility of barley to PM Blumeria graminis f. sp. hordei compared with control lines. It was found that silencing of HvGsl6 does not lead to activation of the SA-dependent defense pathway. Whether reduced susceptibility to powdery mildew species in pmr4/gsl5 mutants is a plant species-specific phenomenon remains to be investigated. We have not been able to check for presence of callose at the cell wall near the sites of fungal penetration in the CRISPR mutants, although we would expect absence or a lower level of callose compared to wild-type plants, depending on redundancy of callose synthase genes. In order to verify whether the PM resistance observed in the tomato pmr4 mutants is callose-independent, quantification of callose deposition should be included in future experiments with these mutants after PM attack.
In tomato an additional possible ortholog of PMR4, designated SlPMR4-h2 (Solyc02g078230), has been found [21]. This gene is the closest tomato ortholog to Arabidopsis gene At4g04970 [21], also known as GSL1/CalS11. Both Solyc07g053980 (GSL5/CalS12-like) and Solyc02g078230 (GSL1/CalS11-like) are reported to function in callose formation during pathogen infection [40]. It would be interesting to investigate whether these genes show functional redundancy in relation to PM resistance/susceptibility, and thus whether knocking out both genes simultaneously would result in higher resistance to PM. However, GSL1 and GSL5 also play redundant roles in plant development [15] . In Arabidopsis, a gsl5 mutant with one mutant allele of GSL1 is severely stunted and shows highly reduced fertility, and double mutants could not be obtained [15].
Perspectives for slpmr4 mutants in breeding for resistance against PM
We have transformed susceptible tomato cultivar Moneymaker with the SlPMR4 CRISPR construct and observed reduced susceptibility in the obtained mutants associated with the activation of the SA signaling pathway. It would be interesting to know what effect the slpmr4 mutation would have in different genetic backgrounds, and especially in resistant tomato backgrounds, with the aim to pyramid different resistance genes. As resistance conferred by the pmr4 mutation is associated with an elevated SA pathway defense response we expect that PM resistance in tomato lines carrying Ol-4 or Ol-6 would not be influenced by mutation of the SlPMR4 gene. Ol-4 and Ol-6 encode NB-LRR type resistance genes [41] and lines containing these genes show a fast HR after PM infection. This strong resistance is not expected to become even stronger because of further elevation of SA levels caused by the pmr4 mutation. Resistance genes Ol-1, Ol-3 and Ol-5 are not cloned yet, but lines containing these genes show a slow HR response. Possibly, combining one of these genes with the pmr4 mutation might result in a stronger or faster HR response. PM resistance conferred by the recessive gene ol-2, containing a mutation in the SlMlo1 gene, is associated with formation of papillae and increased callose deposition. It would be interesting to analyze the PM resistance level in double mutants slmlo1 slpmr4. The resistance conferred by ol-2 might be compromised when the callose synthase gene SlPMR4 is mutated and there is no redundancy with another callose synthase gene (e.g. SlPMR4-h2). However, in Arabidopsis the atmlo2 atpmr4 double mutant displayed the same level of resistance to PM G. orontii as the single atmlo2 mutant [42]. Therefore, the atmlo2 resistance was independent of PMR4-mediated callose deposition [42]. Whether the same holds true in tomato and the slmlo1 slpmr4 double mutant would still show the same level of resistance as the single slmlo1 (ol-2) mutant remains to be tested.
Conclusions
The use of S-genes in plant breeding stands as a promising alternative due to its durable and broad-spectrum characteristics. In this study, we used CRISPR/Cas-9 targeted mutagenesis to knock-out the S-gene PMR4 in tomato. We characterized five different mutation events and confirmed the reduced susceptibility of the mutant lines against On. Our study demonstrates the efficiency and versatility of the CRISPR/Cas9 system as a powerful tool to study and characterize S-genes.
Methods
Design of gRNAs and transformation
The full-length CDS of the tomato PMR4 homolog (Solyc07g053980) was retrieved from the Sol Genomics Network database [43]. Four single guide RNAs (sgRNAs) targeting the gene were selected using the guidelines described by Liang et al. [35] (Supplementary Table 1). A first list of gRNAs was generated using the CC-Top CRISPR/Cas9 Target Prediction Tool [44]. The G + C content of the sgRNAs was calculated using the ENDMEMO webtool (http://www.endmemo.com/bio/gc.php). The folding of the gRNAs was predicted using the Mfold web server [45]. Additionally, the activity of the gRNAs was predicted using the sgRNA scorer [46]. Four sgRNAs were selected (sgRNA1:TTAAAGCAGTCCCATACTCG, sgRNA6: GTACTGCCCCACACTCTGCG, sgRNA7: GCCAAGGTTGCCAGTGGCAA, and sgRNA8: GGATATCAGAGAAGGATCAG) for transformation. The analysis of the location of the sgRNAs and topology of the predicted protein was made using a set of twelve plasmids obtained from Addgene was used to build the construct used for transformation: pICH86966 (as template for amplification); pICSL01009 (as level 0 plasmid); pICH47751, pICH47761, pICH47772, pICH47781 and pICH47732 (as level 1 plasmids); pICH41766, pICH41780 and pICH41822 (as linkers); and pAGM4723 (as level 2 binary vector) (Supplementary figure3). The plasmids were cloned using E. coli DH5α and transformed to Agrobacterium strain AGL1. Susceptible tomato cultivar Moneymaker (from WUR-Plant Breeding seed collection) was used for transformation according to the method described in [47] according to Dutch legislation under GMO licence 01–135.
PCR-based selection of slpmr4 mutants and characterization of mutation events
Selection of plants carrying deletions in SlPMR4 was done by analyses of PCR products flanking the sgRNA target regions. PCR using primers Fw519 (5′- TGGTGCTCTTTTCTCGGTCT-3′) and Rv1925 (5′-CAACTGCTCTTCTGGCATCA-3′) yields a 1407-bp product flanking the sgRNA6 target for the WT allele, and PCR with primers Fw2969 (5′-GCGAATGCGTAGAGAAGGAA-3′) and Rv4230 (5′-CCCCACTAAGTGCCAGGTAA-3′) yields a 1262-bp PCR product flanking sgRNAs8, 1 and 7 for the WT allele. Smaller sizes of the amplified fragments in transgenic plants compared to the WT allele indicated deletions in the targeted region. The sgRNA6 target site together with the PAM site contains the recognition sequence for restriction enzyme XcmI. Fw519 + Rv1925 PCR products were digested with this enzyme (New England Biolabs) yielding 1069-bp and 338-bp fragments for the WT SlPMR4 allele. The PCR products were sequenced to further characterize the mutation events. Primary transformants (T1) carrying mutant alleles were selected using these methods and selfed to produce T2 progeny. Homozygous plants from two T2 bi-allelic lines (TV171030 and TV171033) were selected for use in the disease assay. Homozygous mutant T2 plants derived from other mono-or bi-allelic T1 transformants were selected and selfed to obtain T3 progeny. These included TV161212-U, TV161212-L, TV171009-U, TV171009-L and TV171010 (where U stands for upper band of PCR products in the agarose gel, and L for lower band). Plants from the individual T2 plants and T3 mutant lines (TV171367, TV171368, TV171359, TV171365, TV171366, TV171355, TV171358, TV171356, TV171370 and TV171371) derived from the selfing of previously selected T2 plants, were also tested in the disease assay.
Off-target analysis
The program Cas-OFFinder [34] at http://www.rgenome.net/cas-offinder/ was used to check for possible off-targets of the four sgRNAs of SlPMR4. Mismatch number was set at 3 or less. To analyze possible off-target mutations in gene Solyc02g078230 (SlPMR4-h2) a PCR was performed using primers PMR4_h2_Fw1 (5′-AACGTGTTCTTGCCGATCCTC-3′) and PMR4_h2_Rv1 (5′-CAAAGTGGCTGCGAGCATACA-3′), yielding a 765-bp PCR product for the WT SlPMR4-h2 allele. PCR products of slpmr4 mutants were sequenced and compared with WT control sequences.
Disease assay and quantification of relative fungal biomass
Ten plants homozygous for each of the alleles in the T2 lines and ten plants from the homozygous T3 lines were inoculated with the Wageningen University isolate of On by spraying 4 weeks-old plants with a suspension of conidiospores obtained from leaves of infected tomato Moneymaker plants and adjusted to a concentration of 3.5*104 spores per ml. Two transgenic lines (TV161196 and TV161209) obtained from the same CRISPR transformation experiment, but carrying the wild-type allele were used as controls. Inoculated plants were grown at 20 ± 2 °C with 70 ± 15% relative humidity and day length of 16 h in a greenhouse of Unifarm of Wageningen University & Research, The Netherlands. Disease index scoring was carried out 10 and 12 days after inoculation. Powdery mildew symptoms were scored visually using a scale from 0 to 3 as described by Bai et al. [7]. For the quantification of relative fungal biomass, plant and fungal genomic DNA was isolated from infected leaves collected at 21 dpi, using an adapted CTAB protocol [48]. 10 ng of DNA were used as a template for amplification. Relative fungal biomass was quantified by real-time PCR using the primer pairs On-Fw/On-Rev amplifying the internal transcribed spacer sequence (ITS) of Oidium neolycopersici [49] and Ef-Fw (5′-GGAACTTGAGAAGGAGCCTAAG-3′)/Ef-Rev (5′-CAACACCAACAGCAACAGTCT-3′) amplifying tomato reference gene Elongation Factor 1α (Ef1α) [50]. The 2-ΔΔCt method [51, 52] was used to calculate the fold-change of the ratio between fungal and tomato gDNA.
Histological analysis
At least two plants of each line were grown together with the plants in the disease assay described above but were infected using a heavier inoculation of 3*105 spores per ml. Infected leaf samples of 4 cm2 were collected 44 h post inoculation, bleached in a 1:3 (v/v) acetic acid/ethanol solution, and stained 48 h later by boiling in 0.005% trypan blue in lactophenol: ethanol (1:2 v/v) solution for 3–5 min and cleared in a nearly saturated aqueous solution of chloral hydrate (5:2 w/v) as described by [53]. The slides were mounted on glass slides with a 1:1 (v/v) glycerol-water solution. Analysis of the slides was done using a Zeiss Axiophot bright field microscope. For quantification of fungal structures and host cell death 50 infection units (O. neolycopersici conidia) were analyzed per genotype, from two slides obtained from two different plants per genotype.
Analysis of PR1 expression
Expression level of the tomato PR1 gene was measured 44 hpi after infection with PM by qPCR. Infected leaf samples were taken from individual homozygous mutant T2 plants representing all five mutation events and control plants (MM and unsuccessful transgenic lines TV161196 and TV161209). Leaf samples were frozen in liquid nitrogen and stored at − 80 °C before being grinded into a fine powder using a pestle and mortar. Total RNA isolation was done using the MagMax™ 96 Total RNA Isolation Kit (Qiagen, Germany). cDNA synthesis was done using the iScript™ cDNA Synthesis Kit (Bio-Rad Laboratories, U.S.A.). 10 ng of cDNA were used as template for the reaction. Expression levels of PR1 were measured using primers SlPR1a_Fw (5′-GTGTCCGAGAGGCCAGACTA-3′) and SlPR1a_Rev (5′-CATTGTTGCAACGAGCCC GA-3′), and compared to the expression of tomato Ef1α reference gene using primers Ef_Fw (5′-ATTGGAAACGGATATGCCCCT-3′) and Ef_Rev (5′-TCCTTACCTGAACGCCTGTCA-3′) [21].
Availability of data and materials
The sequences obtained by Sanger sequencing in this study are available in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) under reference accession numbers MT521499 to MT521504. The data produced for this article are included within the manuscript and additional files, and the raw data are available from the corresponding author on reasonable request.
Abbreviations
- CRISPR:
-
Clustered Regularly Interspaced Short Palindromic Repeats
- Cas:
-
CRISPR-associated systems
- dpi:
-
Days post inoculation
- hpi:
-
Hours post inoculation
- NIL:
-
Near-isogenic line
- On :
-
Oidium neolycopersici
- PAM:
-
Protospacer adjacent motif
- PM:
-
Powdery mildew
- PMR4 :
-
Powdery Mildew Resistance 4
- PTTC:
-
Premature translation termination codon
- R gene:
-
Resistance gene
- SA:
-
Salicylic acid
- sgRNA:
-
Single-guide RNA
- S gene:
-
Susceptibility gene
- WT:
-
Wild type
References
Jones H, Whipps JM, Gurr SJ. The tomato powdery mildew fungus Oidium neolycopersici. Mol Plant Pathol. 2001;2:303–9.
Lebeda A, Mieslerová B, Petřivalský M, Luhová L, Špundová M, Sedlářová M, Nožková-Hlaváčková V, Pink DAC. Resistance mechanisms of wild tomato germplasm to infection of Oidium neolycopersici. Eur J Plant Pathol. 2014;138:569–96.
Seifi A, Gao D, Zheng Z, Pavan S, Faino L, Visser RGF, Wolters AMA, Bai Y. Genetics and molecular mechanisms of resistance to powdery mildews in tomato (Solanum lycopersicum) and its wild relatives. Eur J Plant Pathol. 2014;138:641–65.
Bai Y, van der Hulst R, Bonnema G, Marcel TC, Meijer-Dekens F, Niks RE, Lindhout P. Tomato defense to Oidium neolycopersici: dominant Ol genes confer isolate-dependent resistance via a different mechanism than recessive ol-2. Mol Plant-Microbe Interact. 2005;18:354–62.
Huang CC, Groot T, Meijer-Dekens F, Niks RE, Lindhout P. The resistance to powdery mildew (Oidium lycopersicum) in Lycopersicon species is mainly associated with hypersensitive response. Eur J Plant Pathol. 1998;104:399–407.
Li C, Bonnema G, Che D, Dong L, Lindhout P, Visser R, Bai Y. Biochemical and molecular mechanisms involved in monogenic resistance responses to tomato powdery mildew. Mol Plant-Microbe Interact. 2007;20:1161–72.
Bai Y, Pavan S, Zheng Z, Zappel NF, Reinstädler A, Lotti C, De Giovanni C, Ricciardi L, Lindhout P, Visser R, Theres K, Panstruga R. Naturally occurring broad-spectrum powdery mildew resistance in a central american tomato accession is caused by loss of Mlo function. Mol Plant-Microbe Interact. 2008;21:30–9.
Kusch S, Panstruga R. mlo-based resistance: an apparently universal “weapon” to defeat powdery mildew disease. Mol Plant-Microbe Interact. 2017;30:179–89.
van Schie CCN, Takken FLW. Susceptibility genes 101: how to be a good host. Annu Rev Phytopathol. 2014;52:551–81.
Pavan S, Jacobsen E, Visser RGF, Bai Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol Breed. 2010;25:1–12.
Gawehns F, Cornelissen BJC, Takken FLW. The potential of effector-target genes in breeding for plant innate immunity. Microb Biotechnol. 2013;6:223–9.
Vogel J, Somerville S. Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc Natl Acad Sci U S A. 2000;97:1897–902.
Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell. 2003;15:2503–13.
Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC. Loss of a callose synthase results in salicylic acid – dependent disease resistance. Science. 2003;301:969–72.
Enns LC, Kanaoka MM, Torii KU, Comai L, Okada K, Cleland RE. Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol Biol. 2005;58:333–49.
Chen X-Y, Kim J-Y. Callose synthesis in higher plants. Plant Signal Behav. 2009;4:489–92.
Schneider R, Hanak T, Persson S, Voigt CA. Cellulose and callose synthesis and organization in focus, what’s new? Curr Op Plant Biol. 2016;34:9–16.
Ellinger D, Naumann M, Falter C, Zwikowics C, Jamrow T, Manisseri C, Somerville SC, Voigt CA. Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol. 2013;161:1433–44.
Eggert D, Naumann M, Reimer R, Voigt CA. Nanoscale glucan polymer network causes pathogen resistance. Sci Rep. 2014;4:4159.
Eichmann R, Hückelhoven R. Accomodation of powdery mildew fungi in intact plant cells. J Plant Physiol. 2008;165:5–18.
Huibers RP, Loonen AEHM, Gao D, Van den Ackerveken G, Visser RGF, Bai Y. Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS One. 2013;8:e67467.
Langner T, Kamoun S, Belhaj K. CRISPR crops: plant genome editing toward disease resistance. Annu Rev Phytopathol. 2018;56:479–512.
Boettcher M, McManus MT. Choosing the right tool for the job: RNAi, TALEN or CRISPR. Mol Cell. 2015;58:575–85.
Housden BE, Perrimon N. Comparing CRISPR and RNAi-based screening technologies. Nat Biotechnol. 2016;34:621–3.
Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep. 2017;7:482.
Zhang Y, Bai Y, Wu G, Zou S, Chen Y, Gao C, Tang D. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017;91:714–24.
Peng A, Chen S, Lei T, Xu L, He Y, Wu L, Yao L, Zou X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J. 2017;15:1509–19.
Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One. 2016;11(4):e0154027.
Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci. 2015;112(11):3570–5.
Hashimoto R, Ueta R, Abe C, Osakabe Y, Osakabe K. Efficient multiplex genome editing induces precise, and self-ligated type mutations in tomato plants. Front Plant Sci. 2018;9:916.
Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–6.
Okada H, Abe M, Asakawa-Minemura M, Hirata A, Qadota H, Morishita K, Ohnuki S, Nogami S, Ohya Y. Multiple functional domains of the yeast 1,3-β-glucan synthase subunit Fks1p revealed by quantitative phenotypic analysis of temperature-sensitive mutants. Genetics. 2010;184:1013–24.
Inoue SB, Takewaki N, Takasuka T, Mio T, Adachi M, Fujii Y, Miyamoto C, Arisawa M, Furuichi Y, Watanabe T. Characterization and gene cloning of 1,3-β-D-glucan synthase from Saccharomyces cerevisiae. Eur J Biochem. 1995;231:845–54.
Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guides endonucleases. Bioinformatics. 2014;30:1473–5.
Liang G, Zhang H, Lou D, Yu D. Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci Rep. 2016;6:21451.
Schmidt C, Pacher M, Puchta H. Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. Plant J. 2019;98:577–89.
Heidel AJ, Clarke JD, Antonovics J, Dong X. Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics. 2004;168:2197–206.
Šašek V, Janda M, Delage E, Puyaubert J, Guivarc’h A, López Maseda E, Dobrev PI, Caius J, Bóka K, Valentová O, Burketová L, Zachowski A, Ruelland E. Constitutive salicylic acid accumulation in pi4kIIIβ1β2 Arabidopsis plants stunts rosette but not root growth. New Phytol. 2014;203:805–16.
Chowdhury J, Schober MS, Shirley NJ, Singh RR, Jacobs AK, Douchkov D, Schweizer P, Fincher GB, Burton RA, Little A. Down-regulation of the glucan synthase-like 6 gene (HvGsl6) in barley leads to decreased callose accumulation and increased cell wall penetration by Blumeria graminis f. sp. hordei. New Phytol. 2016;212:434–43.
Adkar-Purushothama CR, Brosseau C, Giguère T, Sano T, Moffett P, Perreault JP. Small RNA derived from the virulence modulating region of the potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell. 2015;27(8):2178–94.
Seifi A, Kaloshian I, Vossen J, Che D, Bhattarai KK, Fan J, Naher Z, Goverse A, Tjallingii WF, Lindhout P, Visser RGF, Bai Y. Linked, if not the same, Mi-1 homologues confer resistance to tomato powdery mildew and root-knot nematodes. Mol Plant-Microbe Interact. 2011;24:441–50.
Consonni C, Bednarek P, Humphry M, Francocci F, Ferrari S, Harzen A. Ver Loren van Themaat E, Panstruga R. Tryptophan-derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiol. 2010;152:1544–61.
Fernandez-Pozo N, Menda N, Edwards JD, Saha S, Tecle IY, Strickler SR, Bombarely A, Fisher-York T, Pujar A, Foerster H, Yan A, Mueller LA. The Sol genomics network (SGN) - from genotype to phenotype to breeding. Nucleic Acids Res. 2015;43:D1036–41.
Stemmer M, Thumberger T, Del Sol KM, Wittbrodt J, Mateo JL. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One. 2015;10:e0124633.
Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15.
Chari R, Yeo NC, Chavez A, Church GM. SgRNA scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity. ACS synth. Biol. 2017;6:902–4.
McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R. Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep. 1986;5:81–4.
Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–5.
Zheng Z, Appiano M, Pavan S, Bracuto V, Ricciardi L, Visser RGF, Wolters AMA, Bai Y. Genome-wide study of the tomato SlMLO gene family and its functional characterization in response to the powdery mildew fungus Oidium neolycopersici. Front Plant Sci. 2016;7:380.
Løvdal T, Lillo C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem. 2009;387(2):238–42.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–7.
Anker CC, Niks RE. Prehaustorial resistance to the wheat leaf rust fungus, Puccinia triticina, in Triticum monococcum (s.s.). Euphytica. 2001;117:209–15.
Acknowledgements
We thank He Meng for her help with the work in the laboratory and greenhouse during the fungal biomass quantification. We thank Fien Meijer-Dekens for her help taking care of the CRISPR plants and Katharina Hanika for her help making Fig. 4. We thank the anonymous reviewers for helpful comments to improve the manuscript.
Funding
MISM was funded by a fellowship from The National Council for Science and Technology CONACYT, Mexico. This funding body did not play a role in the design of the study or collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
MISM was involved in designing experiments, characterizing, selecting and testing the mutants, interpreting the results and drafting the manuscript. VB designed the transformation constructs, generated, tested and did the initial characterization of the mutants. EK and MA were involved in selecting and characterizing the mutation events, expression analysis, histological analysis and carrying out the disease assays. EJ, RGFV and YB conceived the idea for the experiments. AMAW was involved in interpreting the results. AMAW and RGFV critically reviewed the paper. AMAW and YB edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests. YB is a member of the editorial board of BMC Plant Biology.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Supplementary Document 1.
Alignment of sequences of PCR products of the tomato PMR4 CRISPR mutant alleles. A. Sequence alignment of PCR products obtained by using primers Fw2969 and Rv4230 for wild type Moneymaker (MM) and five PMR4 CRISPR mutation events. Primers and sgRNA1, 7 and 8 are indicated. In red, nucleotides differing form the MM allele are shown. Deletions are indicated by dashes. Event 5 contains a large inversion, indicated in red. B. Plot showing the inversion (blue line) of the sequence between sgRNA8 and sgRNA7 in mutation event 5 compared to MM.
Additional file 2: Supplementary Figure 1.
Alignment of predicted proteins of the tomato PMR4 CRISPR mutant alleles. Protein sequences are based on DNA sequencing data from the fragments amplified by the region flanked by primers Fw2969 and Rv4230.
Additional file 3: Supplementary Figure 2.
Panel of phenotypes upon infection with Oidium neolycopersici. Leaves from wild-type allele-carrying controls and individual plants of the different slpmr4 mutation classes are shown. Heavy fungal sporulation is present on the leaves of the wild-type plants, while less infection is seen on the leaves of the mutant plants.
Additional file 4: Supplementary Figure 3.
Map of the level 2 vector for CRISPR/Cas9 transformation. The NPTII, Cas9, the four sgRNAs and AtU6 promoters are highlighted.
Additional file 5: Supplementary Table 1.
Characteristics of four selected sgRNAs for SlPMR4. sgRNAs were selected using CCTop program. PAM, protospacer adjacent motif.
Additional file 6: Supplementary Table 2.
SlPMR4 sgRNAs off-targets. Off-targets with a maximum of three mismatches were found for sgRNA8 and sgRNA7 with Cas-OFFinder [34]. crRNA, sgRNA sequences. DNA, off-target sequences. Mismatches are indicated in red.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Santillán Martínez, M.I., Bracuto, V., Koseoglou, E. et al. CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol 20, 284 (2020). https://doi.org/10.1186/s12870-020-02497-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-020-02497-y