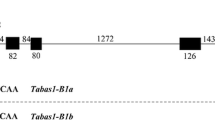Abstract
Background
AFP is a negative regulator of ABA signaling that promotes ABI5 protein degradation and weakens regulation of ABA signaling by targeting upstream genes of ABI5, and TaABI5 gene was seed-specific, and accumulated during wheat grain maturation and dormancy acquisition, which played an important role in seed dormancy; TaAFP has a conserved domain with AFP, so TaAFP may also play an important role in seed dormancy in wheat.
Results
Two allelic variants of TaAFP were identified on chromosome 2BS in common wheat, and designated as TaAFP-B1a and TaAFP-B1b. Sequence analysis showed a 4-bp deletion in the 5’UTR region of TaAFP-B1b compared with TaAFP-B1a. Based on the 4-bp deletion, a co-dominant functional marker of TaAFP-B was developed and designated as AFPB. The genotype generating a 203-bp fragment (TaAFP-B1b) was more resistant to pre-harvest sprouting than the genotype producing a 207-bp fragment (TaAFP-B1a) in a test of 91 white-grained Chinese wheat cultivars and advanced lines. The average germination index(GI) values of TaAFP-B1a and that of TaAFP-B1b were 45.18 and 30.72%, respectively, indicating a significant difference (P < 0.001). Moreover, the 4-bp deletion located in the 5’UTR not only affected the transcription level of TaAFP-B but also affected the mRNA decay, reduced the translation level of GUS and tdTomatoER and GUS activity in wheat leaves of transient expression. The transcript expression and the mRNA half-life value of TaAFP-B1a in developing seeds and mature seeds were much higher than those of TaAFP-B1b.
Conclusion
We identified a 4-bp InDel in the 5’UTR of TaAFP-B, which affected the mRNA transcription level, mRNA decay, translation levels of GUS and tdTomatoER, GUS activity, and was significantly associated with seed dormancy in common wheat. A functional marker was developed and validated based on this InDel.
Similar content being viewed by others
Background
A high level of seed dormancy plays a pivotal role in resistance to pre-harvest sprouting (PHS), providing a mechanism for plants to delay germination until conditions are optimal for survival of the next generation [1, 2]. The balance of abscisic acid (ABA) and gibberellin (GA) levels and sensitivity is a major regulator of dormancy status. The mechanism of ABA sensitivity in seeds has been extensively studied in Arabidopsis. Some genes associated with seed dormancy have been identified as factors in the ABA signaling and ABA synthesis pathway [3,4,5,6]. abi5 is insensitive to an ABA-induced post-germination growth arrest [7], and alters activity of an ABA-inducible late embryo genesis abundant (LEA) gene promoter [8]. ABI5 is mainly expressed in dry seeds, and its expression significantly decreases after germination [9, 10]. Expression of ABI5 is induced strongly by exogenous ABA [4], and is regulated by ABI3, HYL1 and HY5 [11, 12], but is repressed by WRKY2, WRKY40, WRKY18 and WRKY60 [13, 14]. Both abi3 and abi5 mutants were initially recovered by virtue of their ability to germinate in the presence of ABA [7, 15, 16]. The Arabidopsis ABI3 gene is an orthologs of Vp-1 and is required for appropriate ABI5 expression [11, 10, 17]. ABI3 encodes a transcription factor and acts together with ABI5 to govern embryonic gene expression and seed sensitivity to ABA [7, 15, 16].
ABI5 binding protein (AFP), a novel negative regulator of ABA signaling that works by facilitating the degradation of ABI5 [18], was isolated using yeast two-hybrid assays; AFP functions in developing seeds and young seedlings [18]. AFP gene transcription and translation increased during seed development and desiccation, ultimately reaching plateau values in mature seeds [18]. ABI5 acts as a critical factor in maturation, dormancy development of seeds, or the dehydration tolerance of young seedlings of Arabidopsis [19, 20].
In wheat (Tritivum aestivum), many genes or QTL are associated with tolerance to PHS, and molecular markers have been developed based on these genes and QTL [21,22,23,24,25,26]. Several major QTL [27,28,29] associated with PHS tolerance were found and some genes associated with PHS tolerance were cloned [21, 30,31,32,33,34,35,36,37]. TaABI5 genes one of which is the same as TaABF, and TaABF mRNA was seed-specific and accumulated during wheat grain maturation and dormancy acquisition, which played an important role in seed dormancy were isolated in wheat [32]. TaABI5s were expressed in developing grain, roots, and leaves [38]. Three wheat AFP genes (TaAFPs) were isolated, located on the short arms of chromosomes 2A, 2B, and 2D, and designated TaAFP-A, TaAFP-B, and TaAFP-D, respectively [38]. The structure of TaAFP consisted of one intron and two exons, including a nuclear localization domain (119–133 aa of AtAFP) in the middle of the deduced amino acid sequence and an ABI5-binding domain (284–335 aa of AtAFP) in the C-terminal region.
ABI5 has an important function in maturation and dormancy development of seeds in Arabidopsis, while AFP promotes ABI5 protein degradation [18]. Moreover, TaAFP has a conserved domain (60 and 69%) with AtAFP in the region of nuclear localization and the ABI5 binding domain, respectively [38]. Therefore, TaAFP may play an important role in seed dormancy in wheat. The objectives of the present study were to: (1) identify allelic variations at the TaAFP locus in Chinese wheat varieties with different levels of seed dormancy; (2) develop a functional marker for use in marker-assisted selection for PHS tolerance; and (3) characterize the transcription regulation mechanism of TaAFP. The identification of new alleles of TaAFP associated with different seed dormancy could also contribute to our understanding of the mechanisms underlying seed dormancy or PHS tolerance in common wheat.
Results
Sequence analysis of three TaAFP homologs in varieties with different levels of seed dormancy
Full sequences of TaAFP-A, TaAFP-B, and TaAFP-D were cloned using genome-specific primers (Table 1). Six new alleles of TaAFPs were found and named according to the 2005 Supplement of the Wheat Gene Catalogue [39]. In these germplasm, TaAFP-A1a and TaAFP-A1b were on chromosome 2AS; TaAFP-B1a and TaAFP-B1b were on chromosome 2BS; and TaAFP-D1a and TaAFP-D1b were on chromosome 2DS. TaAFP-A was amplified and sequenced with three primer sets TaAFP-AF1/R1, TaAFP-AF2/R2, and TaAFP-AF3/R3 from 10 varieties mentioned above. Two new alleles of TaAFP-A were found and designated TaAFP-A1a and TaAFP-A1b.
Compared with the TaAFP-A (AB360911) [38], 6 SNPs were found in the full sequence of TaAFP-A1a: an A to G transversion was observed at − 197 bp in the 5’UTR; 2 SNPs were located in exons (i.e. C to T at position 94 bp in the first exon, and G to A at position 1523 bp in the second exon) that cause changes of amino acids Gla to Val and Glu to Lys, respectively); and the other 3 SNPs (i.e. G to T, T to A, and G to A) were present in the introns at positions of 529, 717, and 980 bp, respectively. For TaAFP-A1b, 4 SNPs were found: a C to T transversion at 94 bp in the first exon causeing a change of amino acid from Gla to Val; and the other 3 SNPs (i.e. T to A, C to A, and C to A) were located in the introns at positions of 717, 764, and 976 bp, respectively (Additional file 1: Figure S1).
TaAFP-B was amplified with the primer sets TaAFP-BF1/R1, TaAFP-BF2/R2, and TaAFP-BF3/R3. Two alleles of TaAFP-B were found, designated TaAFP-B1a and TaAFP-B1b. Compared with the TaAFP-B gene (GenBank accession AB360912) [38], TaAFP-B1a had 8 SNPs and 2 insertions. Four SNPs (i.e. G to A, G to A, T to C, and G to T at positions − 199 bp, − 152 bp, − 27 bp, and − 26 bp, respectively) were found in the 5’UTR; 2 SNPs (i.e. A to G at position 203 bp that causes change of amino acid from Tyr to Cys, and T to C at 1238 bp that is a synonymous mutation) were found in the first and second exon, respectively; 2 SNPs (i.e. T to C and G to A at position 574 bp and 1102 bp) were observed in introns; and the 2 insertions (G insertion and CT insertion at positions − 45 bp and − 25 bp) were present in the 5’UTR.
TaAFP-B1b had 3 SNPs, 1 insertion, and 1 deletion. One SNP was a C to T at position − 199 bp was found in the 5’UTR; another SNP was A to G at position 1252 bp that causes a change of amino acid from Glu to Gly; and the final SNP was T to C at position 574 bp was identified in an intron; there was a G insertion at position − 45 bp and CT deletion at position − 25 bp were found in the 5’UTR. In addition, a polymorphic fragment was detected in the 10 varieties with different levels of seed dormancy amplified with primer set TaAFP-BF1/R1. An 830-bp fragment was amplified in genotypes of TaAFP-B1a, whereas an 826-bp fragment was generated in genotype of TaAFP-B1b. Compared with the TaAFP-B1a genotype, a 4-bp deletion (CTCT) in the 5’UTR was present in TaAFP-B1b (Additional file 2: Figure S2).
The full sequence of TaAFP-D was amplified with the genome-specific primer sets TaAFP-DF1/R1, TaAFP-DF2/R2, and TaAFP-DF3/R3. Two new alleles of TaAFP-D were found and designated TaAFP-D1a and TaAFP-D1b. Compared with the TaAFP-D gene (GenBank accession AB360913) [38], 3 SNPs were found in TaAFP-D1a: 2 SNPs were located in the second exon (i.e. A to G transversions at positions 1502 and 1530 bp, resulting in changes of amino acids Ile to Met and Ser to Gly, respectively), and another SNP was located in the intron at 1273 bp. Five SNPs were found in the full sequence of TaAFP-D1b: 2 SNPs (i.e. A to G transition at positions 1502 and 1530 bp) were located in the second exon leading to change of amino acids Ile to Met and Ser to Gly, respectively); and the other 3 SNPs (i.e. G to A, T to C, and G to C) were detected in the intron at positions of 742, 995, and 1071 bp (Additional file 3: Figure S3).
Development and validation of the STS marker AFPB for PHS tolerance
Based on the sequence analysis, an STS marker of TaAFP-B, designated AFPB and located in the 5’UTR, was developed and used for association analysis with 91 Chinese varieties and advanced lines. Among the 91 varieties and lines tested, 23 possessed the allele TaAFP-B1a with a 207-bp fragment, whereas 68 had TaAFP-B1b with a 203-bp fragment (Additional file 4: Table S1 and Fig. 1). The GI values of the 91 varieties were consistent over the 2 years (r = 0.631, P < 0.01), with mean values and standard deviations of 0.345 ± 0.17 in 2006 and 0.343 ± 0.16 in 2007. Analysis of variance indicated significant differences (P < 0.001) between the two genotypes. The genotype TaAFP-B1a with a 207 bp fragment was more susceptible to PHS with an average GI value of 0.452, compared with TaAFP-B1b with an average GI value of 0.307, exhibiting a significant association of TaAFP-B with PHS tolerance.
PCR fragments amplified with the primer set TaAFP-BF1/R1 from 10 wheat cultivars and lines. 1: Jimai 19 (TaAFP-B1a); 2: Jinan 16 (TaAFP-B1b); 3: Zhou 8425B (TaAFP-B1a); 4: Yangxiaomai (TaAFP-B1b); 5: Zhoumai 16 (TaAFP-B1a); 6: Yanfu 188 (TaAFP-B1a); 7: Langzhongbaimaizi (TaAFP-B1b); 8: Wanxianbaimaizi (TaAFP-B1b); 9: Yongchuanbaimaizi (TaAFP-B1a); 10: Xiaobaiyuhua (TaAFP-B1b)
Expression of the TaAFP-B gene in Zhou 8425B and Wanxianbaimaizi at different developmental stages
To evaluate the potential influence of different alleles and characterize the expression patterns of TaAFP-B1a and TaAFP-B1b in different cultivars, the expression patterns of Zhou 8425B (TaAFP-B1a) and Wanxianbaimaizi (TaAFP-B1b) were determined using real-time Quantitative PCR (RT-qPCR) analysis. The transcript expression levels of TaAFP-B1a were higher than that of TaAFP-B1b in seeds at 10 days after pollination (DAP), 20 DAP, 30 DAP, 40 DAP, and dry mature seeds immersed in water for 24 h (Fig. 2). In addition, the transcript expression levels of TaAFP-B1a and TaAFP-B1b had a trend of first increasing and then decreasing in seeds of different developmental stages, reaching the highest level at 20 DAP and the lowest at 40 DAP (Fig. 2). No transcript expression of TaAFP-B1a and TaAFP-B1b was detected in dry seeds. The results illustrated that allelic variation in the 5’UTR of TaAFP-B affected the transcript expression level in seeds at 10 DAP, 20 DAP, 30 DAP, and 40 DAP and in dry mature seeds immersed in water for 24 h.
Effect of the 4-bp deletion in the 5’UTR on the mRNA decay of TaAFP-B
The results showed that TaAFP-B mRNA half-life (t1/2) of Zhou 8425B with the allele TaAFP-B1a was 144.38 min, 154.00 min, and 115.50 min, in flag leaves, dry mature seeds, and seeds at 20 DAP, respectively, whereas those of Wanxianbaimaizi were 39.60 min, 46.20 min, and 33.32 min, respectively (Fig. 3). This indicated that TaAFP-B mRNA of Zhou 8425B with TaAFP-B1a was more stable than that of Wanxianbaimaizi with TaAFP-B1b.
The mRNA expression level of genotypes of Zhou 8425B (TaAFP-B1a) and Wanxianbaimaizi (TaAFP-B1b) treated with Cordycepin for 0 min and 60 min in dry seeds immersed in water for 24 h, 20 DAP seeds, and flag leaves. a Dry seeds immersed in water for 24 h: the half-life of TaAFP-B is 154.00 min in Zhou 8425B and 46.20 min in Wanxianbaimaizi; b 20 DAP seeds: the half-life of TaAFP-B is 115.50 min in Zhou 8425B and 33.32 min in Wanxianbaimaizi; c Flag leaves: the half-life of TaAFP-B is 144.38 min in Zhou 8425B and 39.60 min in Wanxianbaimai
The mRNA patterns of TaAFPs and TaABI5 at different seed developmental stages
RT-qPCR showed that TaAFP-B had the highest transcription level of the four genes: TaAFP-A, TaAFP-B, TaAFP-D, and TaABI5 at 10 DAP, 20 DAP, 30 DAP, and 40 DAP, respectively, in two genotypes of Zhou 8425B and Wanxianbaimaizi. Transcription levels of these four genes also had the general trend of gradually increasing and then decreasing during seed development. At the same time, TaAFP-B had a higher transcriptional level in Zhou 8425B than in Wanxianbaimaizi at each observation time point (Fig. 4). The most abundant TaAFP-B transcript levels in Zhou 8425B and Wanxianbaimaizi were detected at 20 DAP, while the highest transcriptional level of TaABI5 occurred at 20 DAP in Zhou 8425B and at 30 DAP in Wanxianbaimaizi (Fig. 4). These results indicated that the time of the highest transcription level of TaABI5 was latter than that of TaAFP-B in Wanxianbaimaizi; this might be another reason why Wanxianbaimaizi has a higher capacity of seed dormancy than Zhou 8425B.
Transient expression of tdTomatoER and GUS-tagged pITV1-TaAFP-Ba/bF in wheat leaves
The plant transient expression vectors pITV1-TaAFP-BaF, pITV1-TaAFP-BbF, and pITV1#1534 (control) were separately transformed into wheat leaves of Zhou 8425B using biolistic bombardment. The fluorescence intensity of tdTomatoER gene expression in leaves of wheat were observed with a confocal laser scanning microscope after the transformation for 12–18 h in a dark environment, then the GUS expression was observed with an optical microscope after treatment with dye and decoloring solution of GUS. The results showed that the fluorescence intensity of tdTomatoER gene expression and in the leaves was in the following order: pITV1-TaAFP-BaF > pITV1-TaAFP-BbF > pITV1#1534 (Fig. 5a, b, and c). In addition, GUS gene expression showed the same trend with the fluorescence intensity of tdTomatoER gene expression (Fig. 5: d, e, and f). These results indicated that the 4 bp deletion in the 5’UTR of TaAFP-B reduced the translation level of tdTomatoER and GUS.
Transient experssion of tdTomatoER and GUS-Tagged TaAFP-Ba/bF in wheat leaves by confocal laser scanning microscope and optical microscope. a The expression of red fluorescence tdTomatoER in leaves of wheat by transforming TaAFP-BaF gene, Meanintensity: 5714.24, meandensity: 3.96; b The expression of red fluorescence tdTomatoER in leaves of wheat by transforming TaAFP-BbF gene, meanintensity: 101.22, meandensity: 3.43; c The expression of red fluorescence tdTomatoER in leaves of wheat in control, meanintensity: 43.14; meandensity: 3.12; d The expression of GUS in wheat leaves by transforming TaAFP-BaF gene; e The expression of GUS in wheat leaves by transforming TaAFP-BbF gene; f The expression of GUS in wheat leaves in control
Quantitative analysis of TaAFP-Ba/bF activity
To investigate the quantitative of TaAFP-Ba/bF sequence of promoter activity, total proteins were extracted from wheat leaves of transient expression. The GUS activity was determined by fluorometric assays. The GUS expression level of TaAFP-BaF::GUS and TaAFP-BbF::GUS transient expression leaves were significantly highter than Col (P < 0.01)(Fig. 6). Otherwise, the GUS expression level and activity in TaAFP-BaF::GUS were higher than TaAFP-BbF::GUS (P < 0.01). The results indicated that the 4 bp deletion in the 5’UTR of TaAFP-B decreased the GUS activity.
Discussion
The 800-bp region in the 5’UTR upstream from the start codon ATG of TaAFPs is relatively well conserved [38]. Nevertheless, in the present study, five allelic variants in the 5’UTR of TaAFP-A and TaAFP-B were found in 10 varieties (Additional file 1: Figure S1 and Additional file 2: Figure S2), indicating rich allelic variations in the 5’UTR of TaAFP-A and TaAFP-B in these germplasms. In addition, no alleles with the same to sequences as AB360911, AB360912, or AB360913 [38] were found in these 10 varieties, which may be attributed to the limited varieties sequenced. Furthermore, among the allelic variations present in TaAFP-A, TaAFP-B and TaAFP-D loci, only a CTCT deletion at the TaAFP-B1b locus was associated with seed dormancy in 91 varieties.
Analysis of TaAFP sequence showed some transcription factor binding elements in the region of 800 bp located upstream from the start code ATG, including ABA response elements ABRE, G-box, CACA, AACAA, Dof, RAVI, Myb-type transcription factor elements P, GAmyb, DRE/CRT and elements for cold and dehydration response [38]. Although the 4-bp deletion in the 5’UTR of TaAFP-B1b did not reside in any of the elements mentioned above, it changed the number of adjacent CT repeats from 9 to 7. Previous reports indicated that the number of repeat sequences in the promoter region of a gene could affect the expression level [40, 41]. Further study also showed that the 4-bp (CTCT) deletion changed the mRNA half-life and further transcription level (Figs. 2 and 3). The sequence with 9 CT repeats might increase the stability of mRNA and the expression level of TaAFP-B1a, which could enhance the binding of related transcription factors through changing the secondary structure of the upstream region of TaAFP-B1a during transcription.
Altering mRNA stability under some conditions plays an important role in the dynamic control of gene expression [42]; the rate of mRNA decay is an essential element of post-transcriptional regulation in all organisms, because the stability of mRNA determines how fast the equilibrium level of a new protein will be reached [43]. Thus, the half-life of mRNA will influence the stochastic fluctuation in the production rate of the corresponding protein [44]. mRNA transcripts are protected from degradation by exoribonuclease by 5′ capping and 3′ poly A structures [45]. In this study, compared with the sequence of TaAFP-B1a, a 4-bp deletion (CTCT) located at − 25 bp of the 5’UTR was found in TaAFP-B1b, and the mRNA half-life values of TaAFP-B1b and TaAFP-B1a were significantly different. More stable mRNA and a higher transcriptional level existed in TaAFP-B1a genotype.
The mRNA degradation data (Fig. 4) in this study suggest that the 4-bp deletion (CTCT) in the 5’UTR of TaAFP-B1b forms a “hot spot” for degradation by endogenous ribonucleases, whereas the region of TaAFP-B1a with “CTCT” sequence might be engineered to be more stable, leading to increased mRNA half-life. Consequently, more protein production and higher expression levels of tdTomatoER and GUS were observed in pITV1-TaAFP-BaF compared to pITV1-TaAFP-BbF, and higher GUS activity were detected in TaAFP-BaF::GUS transient expression wheat leaves than TaAFP-BbF::GUS, which indicated that the difference of the 4-bp deletion (CTCT) affected not only mRNA decay and transcription expression level, but also the translation expression level of its downstream gene.
Seed dormancy is a complex quantitative trait. In wheat, ABA signaling is the main factor for seed dormancy [46, 47]. TaAFP is a negative regulator in seed dormancy in wheat. In the present study, an STS marker AFPB associated with seed dormancy in Chinese wheat cultivars with different GI values was developed. In this set of germplasms, there are significant differences in GI between the TaAFP-B1a and TaAFP-B1b genotypes (P = 0.0002). Several STS markers like Vp1A3, Vp1B3, TaSdr and Tamyb10D were associated with seed dormancy in wheat [21, 22, 25]. The TaVp-1, TaAFP, and TaSdr genes were involved in the mechanism of embryo-imposed dormancy, while Tamyb10 was involved in the mechanism of coat-imposed dormancy. It is better to obtain highly efficient marker-assisted selection for PHS-resistant varieties by combining both the embryo-imposed and coat-imposed dormancy.
Conclusions
In this study, two allelic variations of TaAFP-B were identified, including TaAFP-B1a and TaAFP-B1b. TaAFP-B had a 4-bp InDel in the 5’UTR, which affected the mRNA stability, mRNA transcription expression level, translation expression level of tdTomatoER and GUS, and GUS activity and was significantly associated with PHS tolerance in common wheat. Based on the 4-bp InDel, a functional marker was developed and validated.
Materials and methods
Plant materials
Ten wheat varieties were used for cloning of TaAFP-A, TaAFP-B, and TaAFP-D, including five PHS-resistant varieties (Xiaoyan6, Langzhongbaimaizi, Wanxianbaimaizi, Yangxiaomai, and Xiaobaiyuhua, with GI values of 0.137, 0.084, 0.076, 0.075, and 0.04, respectively) and five PHS-susceptible varieties (Yongchuanbaimaizi, Xinong 979, Zhoumai 16, Zhou 8425B, and Jinan 16, with GI values of 0.232, 0.327, 0.494, 0.560, and 0.616, respectively). Ninety-one Chinese wheat varieties and advanced lines with different PHS tolerances, from the China Autumn-sown Wheat Region (CAWR) representing more than 85% of wheat production areas in China, were used for association analysis of TaAFP-B allelic variations and PHS tolerance. The GI was determined based on the average date across two cropping seasons at two locations in Beijing and Anyang (Henan) in 2005–2006 and 2006–2007 (Additional file 4: Table S1). Grains of Zhou 8425B and Wanxianbaimaizi planted in Hohhot in Inner Mongolia after seeds vernalization in 2016, and collected the spikes at 10, 20, 30, and 40 DAP, and at maturity were frozen in liquid nitrogen, and stored at − 80 °C for analysis of mRNA transcription. Gene gun transformation was conducted during the period when there was one leaf and one terminal bud of wheat.
Primer design
Nine gene-specific primers, TaAFP-AF1/R1, TaAFP-AF2/R2, TaAFP-AF3/R3, TaAFP-BF1/R1, TaAFP-BF2/R2, TaAFP-BF3/R3, TaAFP-DF1/R1, TaAFP-DF2/R2, and TaAFP-DF3/R3, were used to clone TaAFP-A, TaAFP-B, and TaAFP-D genes (Table 1). Another primer AFPBF/R was a specific marker used for determining the allelic variants of TaAFP-B detected by 10% denaturing polyacrylamide gels. The primer Q-TaAFP-BF/R, Q-TaAFP-AF/R, Q-TaAFP-DF/R, and Q-ABI5F/R were designed to perform analysis of mRNA expression levels of TaAFP-A, TaAFP-B, TaAFP-D, and TaABI5. The wheat ACTIN gene was used as an internal control and included in each reaction in order to normalize the expression levels of TaAFP-A, TaAFP-D, and TaABI5 genes in Zhou 8425B and Wanxianbaimaizi, and the expected PCR product was 410-bp in length (Table 1).
DNA and RNA extraction
Genomic DNA was extracted from 3 g of seedlings grown in the dark at 25 °C for 7 days by the CTAB method [48]. Total RNA was extracted from five whole grains at different developmental stages with the TransZol Plant Kit (TransGenBiotech).
PCR amplification and RT-qPCR analysis
PCR reactions for gene cloning and molecular marker tests were performed in an MJ Research PTC-200 thermal cycler in a total volume of 15 μl, including 1.5 μl of 10 × PCR buffer, 1.2 μl of 2.5 μM dNTP each, 4 pmol of each primer, 0.75 U of LaTaq polymerase (TaKaRa), and 500 ng of template DNA, then up to 15 μl with ddH2O. PCR amplification were 94 °C for 5 min, followed by 35 cycles of 94 °C for 45 s, 57 °C–65 °C for 45 s, and 72 °C for 45 s, with a final extension of 72 °C for 10 min. Amplified PCR fragments were separated on 1.5% agarose gel with the nucleic acid dye Gelview (TaKaRa).
The cDNA was synthesized from 5 μg of total RNA using M-MLV reverse transcriptase (TaKaRa) with random hexamer primer Oligo (dT)19 according to the manufacturer’s instructions. RT-qPCR reactions were performed in a LightCycler®480 Real Time PCR System following the introduction book of SYRB® Premix Ex Taq™ II (TLiRnaseH Plus). PCR cycling was performed at 94 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, 55 °C − 66 °C for 30 s and 72 °C for 30 s, with a final extension at 72 °C for 10 min. Then, the expression level of the TaAFP-B gene was estimated based on the size of the 102-bp fragment amplified with Q-TaAFP-BF/R primers designed in the exon region. The gene transcriptional expression analysis of TaAFP and TaABI5 were also estimated also with RT-qPCR; which primers showed in Table 1.
mRNA half-life assay
The TaAFP-B mRNA half-life (t1/2) was determined in flag leaves at the heading stage, dry mature seeds immersed in water for 24 h and seeds at 20 DAP of cultivars Zhou 8425B (TaAFP-B1a) and Wanxianbaimaizi (TaAFP-B1b), treated with 200 ng/mL Cordycepin (Sigma-Genosys, USA) for 0 min and 60 mins, respectively. The mRNA half-life (t1/2) was calculated based on the formula: T(1/2) = 0.693/kd [49], where the denominator kd is a gradient constant determined by the value of Ct in the real-time RT-PCR reaction.
DNA sequencing
The PCR products were sequenced from both strands by the Beijing Genomics Institute (http://www.genomics.cn). Sequence analysis and characterization were performed using the software DNAMAN (http://www.lynnon.com).
Statistical analysis
Analysis of variance was conducted by PROC MIXED in the Statistical Analysis System (SAS Institute, 8.0) with genotype clusters indicated by two types of fragments amplified with the STS marker AFPB, as a categorical variable to derive mean GI value from each cluster and to test significance levels. The genotype clusters were treated as fixed effects, while genotypes nested in clusters and years were treated as random effects. Pearson’s linear correlation coefficients for GI between years were obtained by SAS PROC CORR.
Plasmid constructions
The plant expression vector of pITV1#1534 was supplied by YP. Xing from Inner Mongolia Agricultural University. Then, the two different fragments (272 bp and 276 bp, the sequences were part of the 5’UTR of TaAFP-B1a and TaAFP-B1b, respectively, with an added sequence of a mini promoter in a 5′ orientation), which differ in the 4 bp deletion (CTCT), were synthesized by General Biosystems (Anhui) Co. Ltd. The confirmed sequences were cloned into NotI and NocI sites of the binary vector pITV1#1534. Finally, two recombinant expression vectors, pITV1-TaAFP-BaF and pITV1-TaAFP-BbF, were constructed successfully (Fig. 7).
Plant transformation and observation
When the leaves of wheat grew to the period of one leaf and one terminal bud, the first leaf was cut off about 4 cm from the tip of the leaf, and soaked in 70% ethanol for 3 min. Then, the leaves were washed with distilled water 3 times, the leaf surface was cleaned, and the leaves were attached to a piece of glass. Finally, plasmid DNA (1 μg/μl) were transferred into leaves by biolistic bombardment (PDS-1000/He system of BioRad) transformation. He pressure of the rupture disc was 1100 psi and vacuum degree was 28 inHg. The bombardment distance was 9 cm [50, 51]. Every experiment had three biological replicates.
The tdTomatoER expression was viewed in whole leaves mounted on glass slides using the Olympus BX-60 of Confocal Laser Scanning Microscope (excitation light 540 nm, emitted light 580 nm). Images were processed with Adobe Photoshop (Mountain View, CA). Then, for observation of GUS expression, the samples were incubated overnight in a solution of 1 mM X-Gluc in 50 mM phosphate buffer (pH 7.0) at 37 °C. After that, tissues were cleared of chlorophyll in 70% ethanol and photographs of whole-mounted tissues were taken using an optical microscope. Finally, all transgenic plants for each construct were analyzed.
Quantitative GUS assay
All transient expression wheat leaves separately converted to pITV1-TaAFP-BaF, pITV1-TaAFP-BbF, and pITV1#1534 as a control by biolistic bombardment were frozen in liquid nitrogen, and stored at − 80 °C prior to the quantitative analysis of GUS activity of the promoter of TaAFP-BaF and TaAFP-BbF, which was expressed as nanomoles 4-MU (4-methylumbelliferone) per minute per microgram protein [52]. For each construct, at least six independent transient expression wheat leaves were analyzed, and three replicates were performed.
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional files. Sequence data used in this manuscript can be found in datebase of NCBI (https://www.ncbi.nlm.nih.gov/) under the following accession numbers: TaAFP-A(AB360911), TaAFP-B(AB360912), TaAFP-D(AB360913).
Abbreviations
- ABA:
-
Abscisic acid
- GA:
-
Gibberellins
- LEA:
-
Late embryo genesis abundant
- PHS:
-
Pre-harvest sprouting
- Vp-1 :
-
Viviparous-1
References
Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. Target genes and regulatory domains of the GAMYB transcription activator in cereal aleurone. Plant J. 1999;17:1–9.
Walker-Simmons M, Sesing J. Temperature effects on embryonic abscisic acid levels during development of wheat grain dormancy. Plant Growth Regul. 1990;9:51–6.
Sugimoto K, Takeuchi Y, Ebana K, Miyao A, Hirochika H, Hara N, Ishiyama K, Kobayashi M, Ban Y, Hattori T, Yano M. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc Natl Acad Sci U S A. 2010;107:5792–7.
Oikawa T, Koshioka M, Kojima K, Yoshida H, Kawata M. A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol Biol. 2004;55:687–700.
Liu HY, Yu X, Cui DY, Sun MH, Sun WN, Tang ZC, Kwak SS, Su WA. The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Res. 2007;17:638–49.
Lee Y, Kende H. Expression of beta-expansins is correlated with internodal elongation in deep water rice. Plant Physiol. 2001;127:645–54.
Lopez-Molina L, Chua NH. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:541–7.
Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M. Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J. 2002;30:373–83.
Lopez-Molina L, Mongrand S, Chua NH. A post germination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci U S A. 2001;98:4782–7.
Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609.
Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32:1–12.
Hao CN, Li MX. Role of HY5 in abscisic acid response in seeds and seedlings. Plant Signal Behav. 2008;3:986–8.
Jiang WB, Yu DQ. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009;9:96.
Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, Zhang XF, Zhao R, Sun HL, Liu R, Yu YT, Zhang DP. The mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 2010;22:1909–35.
Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plantarum. 1984;61:377–83.
Finkelstein RR. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutation. Plant. 1994;5:765–71.
Soderman EM, Brocard IM, Lynch TJ, Finkelstein RR. Regulation and function of the Arabidopsis ABA insensitive 4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 2000;124:1752–65.
Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 2003;17:410–8.
Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:15–45.
Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 2006;18:3415–28.
Yang Y, Zhao XL, Xia LQ, Chen XM, Xia XC, Yu Z, He ZH. Development and validation of a Viviparous-1 STS marker for pre-harvest sprouting tolerance in Chinese wheats. Theor Appl Genet. 2007a;115:971–80.
Yang Y, Zhang CL, Liu SX, Sun YQ, Meng JY, Xia LQ. Characterization of the rich haplotypes of Viviparous-1A in Chinese wheats and development of a novel sequence-tagged site marker for pre-harvest sprouting resistance. Mol Breed. 2014;33:75–88.
Zhang YJ, Miao XL, Xia XC, He ZH. Cloning of seed dormancy gene (TaSdr) associated with tolerance to pre-harvest sprouting in common wheat and development of a functional marker. Theor Appl Genet. 2014;127:855–66.
Bi HH, Sun YW, Xiao YG, Xia LQ. Characterization of DFR allelic variations and their associations with pre-harvest sprouting resistance in a set of red-grained Chinese wheat germplasm. Euphytica. 2014;195:197–207.
Wang Y, Wang XL, Meng JY, Zhang YJ, He ZH, Yang Y. Characterization of Tamyb10 allelic variants and development of STS marker for pre-harvest sprouting resistance in Chinese bread wheat. Mol Breed. 2016;36:148.
Rasul G, Humphreys DG, Brûlé-Babel A, McCartney CA, Knox RE, DePauw RM, Somers DJ. Mapping QTLs for pre-harvest sprouting traits in the spring wheat cross ‘RL4452/AC domain’. Euphytica. 2009;168:363–78.
Chen CX, Cai SB, Bai GH. A major QTL controlling seed dormancy and pre-harvest sprouting resistance on chromosome 4A in a Chinese wheat landrace. Mol Breed. 2008;21:351–8.
Osa M, Kato K, Mori M, Shindo C, Torada A, Miura H. Mapping QTLs for seed dormancy and the Vp1 homologue on chromosome 3A in wheat. Theor Appl Genet. 2003;106:1491–6.
Liu SB, Bai GH. Dissection and fine mapping of a major QTL for pre-harvest sprouting resistance in white wheat rio Blanco. Theor Appl Genet. 2010;121:1395–404.
Anderberg RJ, Walker-Simmons MK. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc Natl Acad Sci U S A. 1992;89(21):10183–7.
Holappa LD, Walker-Simmons MK, Ho THD, Riechers DE, Beckles DM, Jones RL. A Triticum tauschii protein kinase related to wheat PKABA1 is associated with ABA signaling and is distributed between the nucleus and cytosol. J Cereal Sci. 2005;41(3):333–46.
Johson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element binding factor, TaABF and phosphorylates TaABFP eptide sequences. Plant Physiol. 2002;130:837–46.
McKibbin RS, Wilkinson MD, Bailey PC, Flintham JE, Andrew LM, Lazzeri PA, Gale MD, Lenton JR, Holdsworth MJ. Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proc Natl Acad Sci U S A. 2002;99:10203–8.
Zhang CL, He XY, He ZH, Wang LH, Xia XC. Cloning of TaCYP707A1 gene that encodes ABA 8′-hydroxylase in common wheat (Triticum aestivum L.). Agric Sci China. 2009;8:902–9.
Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, Mori M, Kawaura K, Ogihara Y, Miura H. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell. 2011;23:3215–29.
Liu SB, Sehgal SK, Li JR, Lin M, Trick HN, Yu JM, Gill BS, Bai GH. Cloning and characterization of a critical regulator for preharvest sprouting in wheat. Genetics. 2013;195:263–73.
Ashikawa I, Abe F, Nakamura S. Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci. 2010;179:536–42.
Ohnishi N, Himi E, Yamasaki Y, Noda K. Differential expression of three ABA-insensitive five binding protein (AFP)-like genes in wheat. Genes Genet Syst. 2008;83:167–77.
McIntosh RA, Devos KM, Dubcovsky J, Rogers WJ, Morris CF, Appels R, Anderson OD Catalogue of gene symbols for wheat: 2005 supplement. Published online at http://wheat.pw.usda.gov/ggpages/wgc/2005upd.html.
Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, Allan AC. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell. 2009;21:168–83.
Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–6.
Fan J, Yang X, Wang W, Wood WH 3rd, Becker KG, Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Natl Acad Sci U S A. 2002;99:10611–6.
Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–50.
McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci U S A. 1997;94:814–9.
Prescott DM. Cells: principles of molecular structure and function: Jones and Bartlett Publishers; 1988.
Kawakami N, Miyake Y, Noda K. ABA insensitivity and low ABA levels during seed development of non-dormant wheat mutants. J Exp Bot. 1997;48:1415–21.
Walker-Simmons M. ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol. 1987;84:61–6.
Gale KR, Ma W, Zhang W, Rampling L, Hill AS, Appels R, Morris P, Morrel M. Simple high-through put DNA markers for genotyping in wheat. In: Eastwood R et al (eds) 10th Australian wheat breeding assembly proceedings. 2001; pp 26–31.
Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, Averett LM, Zhao H, Davis RE, Sathyamoorthy M, Wahl LM, Harris ED, Mikovits JA, Monks AP, Hollingshead MG, Sausville EA, Staudt LM. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anticancer drug flavopiridol. Genome Biol. 2001;2(10):research 0041.1–0041.11.
Yahiaoui N, Srichumpa P, Dudler R, Keller B. Genome analysis at different ploidy levels allows cloning of the powery mildew resistance gene Pm3b from hexaploid wheat. Plant Journal. 2004;37:528–38.
Xu HH, Liu JJ, Li N, Ma ZQ. Optimization of transient expression system in leaves of wheat. J Triticeae Crops. 2015;35(12):1617–23.
Li J, Xu RF, Qin RY, Ma H. Hao Li, Zhang YP, Li L, Wei PC, Yang JB. Isolation and functional characterization of a novel rice constitutive promoter. Plant Cell Rep. 2014;33:1651–60.
Acknowledgements
Not applicable.
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 31760382), the Science and Technology Project of Henan Province (Grant No. 152102110136 and 172102110076), the Inner Mongolia Science & Technology Plan-Innovation Team (Grant No. 201503004),The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
YF. performed the experiments and wrote the paper. ML, YX and ZW assisted to perform the experiment. XZ, BH and MW planted the experimental materials and identified the GI values. YY and YX designed the experiment and assisted in writing the paper. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
We declare that these experiments comply with the ethical standards in China where they were performed.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. Sequence comparison of two new TaAFP-A alleles of TaAFP-A1a and TaAFP-A1b detected in Chinese germplasm with TaAFP-A (AB360911).SNPs are in bold letters. (DOCX 19 kb)
Additional file 2:
Figure S2. Sequence comparison of three new TaAFP-B alleles of TaAFP-B1a, TaAFP-B1c and TaAFP-B1b detected in Chinese germplasm with TaAFP-B (AB360912). Insertions are underlined, deletions are shadowed, and SNPS are in bold letters. (DOCX 17 kb)
Additional file 3:
Figure S3. Sequence comparison of two new TaAFP-D alleles of TaAFP-D1a and TaAFP-D1b detected in Chinese germplasm with TaAFP-D (AB360913). SNPS are in bold letters. (DOCX 19 kb)
Additional file 4:
Table S1. Polymorphism of the AFPB marker in 91 white-grained cultivars with different levelsof seed dormancy. (DOCX 20 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Feng, Y., Liu, M., Wang, Z. et al. A 4-bp deletion in the 5’UTR of TaAFP-B is associated with seed dormancy in common wheat (Triticum aestivum L.). BMC Plant Biol 19, 349 (2019). https://doi.org/10.1186/s12870-019-1950-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-019-1950-4