Abstract
Background
Orphan nuclear receptor estrogen related receptor β (Esrrb or ERRβ) is well known in stem cells and early embryonic development. However, little is known about its function in cancer.
Method
We investigated the mRNA profile alterations induced by Esrrb expression and its synthetic ligand DY131 in human prostate cancer DU145 cells via RNA-Seq analysis.
Results
We distinguished 67 mRNAs differentially expressed by Esrrb alone. Although DY131 alone did not change any gene, treatment of DY131 in the presence of Esrrb altered 1161 mRNAs. These observations indicated Esrrb had both ligand-independent and ligand-dependent activity. When Esrrb was expressed, DY131 treatment further regulated 15 Esrrb-altered mRNAs. DY131 acted as an antagonist for 11 of 15 mRNAs (wdr52, f13a1, pxdn, spns2, loc100506599, tagln, loc441454, tkel1, sema3f, zcwpw2, sdc2) and as an agonist for 4 of the 15 mRNAs (rarres3, oasl, padi2, ddx60). Gene ontology analyses showed altered genes are related to transcription and translation regulation, cell proliferation and apoptosis regulation, and cellular metabolism.
Conclusion
Our results characterized mRNA profiles in DU145 prostate cancer cells driven by Esrrb expression and Esrrb ligand DY131, and provided multiple markers to characterize Esrrb’s function in Esrrb research.
Similar content being viewed by others
Background
Esrrb encodes nuclear receptor estrogen related receptor β (Esrrb), which belongs to the nuclear receptor family. Esrrb acts as a transcription factor by binding to a specific DNA sequence estrogen related receptor response element (ERRE), which is also known as steroid factor response element (SFRE), or half site estrogen response element [1, 2].
Esrrb, first cloned in 1988, was not intensively studied until recent years. Knocking out of Esrrb was embryonic lethal due to placental malformation [3]. Though early studies showed a very limited range of tissues with positive Esrrb expression, recent studies reported that short form Esrrb alternative splicing isoform had a broad range of expression [4]. Esrrb was found to be a core-reprogramming factor to reprogram Pluripotent Stem Cells (iPSCs) [3–6]. c-myc and klf4 of the OSKM (oct4, sox2, klf4, c-myc) core-reprogramming factors can be replaced by Esrrb [5, 6]. Esrrb was also recently reported to drive sox2 transcription and induce iPSC in a single cell system [7].
Tumorigenesis and tumor progression are related to Esrrb. Esrrb was shown to be down-regulated in prostate cancer epithelium compared to normal prostate tissue [8–10]. Its re-expression in DU145 and LNCaP cells was shown to stimulate tumor suppressor cdkn1a (p21) concentration. Also, Esrrb can inhibit Estrogen Receptor transcriptional activity in uterine endometrial cancer cells and Nrf2-Keap signaling pathway in breast cancer cells [11, 12].
There are a handful of transcriptome-wide expression survey data from Esrrb knockdown in both human iPSCs and mouse embryonic stem cells [13–16]. Known Esrrb controlled genes include klf4, c-myc, cdkn1a and cyp19a1, but Esrrb target genes in cancer cells are still not known.
This manuscript focuses on the discovery of Esrrb ligand-independent and Esrrb ligand-dependent target genes. We performed RNA-Seq analysis to characterize Esrrb regulated mRNAs in a prostate cancer cell line and we found the treatment of DY131 expanded Esrrb’s transcriptional regulation activity to many more genes.
Results
Establishment of the Esrrb stably transfected DU145 cells
Esrrb expression vector or control pcDNA3.1 (Zeo+) vector were transfected into DU145 cells. After 3 weeks of Zeocine selection, we characterized the Esrrb status by reverse transcriptase (RT)-PCR, qPCR and western blot analysis (Fig. 1a–c). Our results showed that Esrrb was successfully expressed in DU145-Esrrb cells. Although RNA-Seq showed that DU145-pc3.1 cells had a very small amount of Esrrb expressed (count per million read <1), the Esrrb concentration is below the detection limit of RT-PCR and western blot. Compared to HEK293 cells, which expressed endogenous Esrrb, overexpression of Esrrb in DU145 cells raised the Esrrb protein concentration to a comparable physiological concentration (Fig. 1b). In addition, our RT-PCR results and RNA-seq results confirmed the estrogen related receptor gamma (Esrrg) was not expressed in DU145 cells. The absence of Esrrg eliminated any possible functional contamination by Esrrg in our Esrrb studies (Fig. 1c).
Characterization of Esrrb-expressing cancer cell line. Esrrb status of two independent replicates of stable transfected control DU145-pc3.1 and DU145-Esrrb cells are tested by a quantitative PCR b Western blot and c reverse transcriptase PCR. a Relative mRNA concentrations of Esrrb were measured by qPCR, Esrrb transcripts concentration were determined by standard curve method and Esrrb concentration were first normalized to the concentration of house keeping gene GAPDH, then normalized to Esrrb/GAPDH ratio of DU145-pc3.1 cells. b Total protein was extracted form HEK293, DU145-Esrrb and control DU145-pc3.1 cells. Protein concentration of Esrrb was determined by western blot using GAPDH as internal control. c RT-PCR was performed on total RNA extracted from HEK293, DU145-esrrb and control DU145-pc3.1 cells. Esrrb was expressed in DU145-Esrrb cells, while Esrrg is not expressed in either DU145-pc3.1 and DU145-Esrrb cells
Esrrb expression alters mRNA profile
To distinguish genes regulated by Esrrb, we performed RNA-Seq analysis on cDNA libraries constructed from two biological replicates of both DU145-pc3.1 and DU145-Esrrb cells. Spearman ranking correlation analysis showed that the expression of Esrrb in DU145 created a distinct transcriptome compared to control DU145-pc3.1 cells (Fig. 2a). We found 67 genes (21 genes up-regulated, 46 genes down-regulated) altered due to Esrrb expression (Fig. 2b; Table 1). Seven genes that are among the most changed genes (zcwpw2, hoxb8, tagln, f13a1, pxdn, aox1, and bmp4, as well as tgfβ as a negative control) were confirmed by qPCR (Fig. 3). Gene ontology (GO) analysis shows that the products of Esrrb driven differentially expressed genes fell into functional categories of regulation of cell development as well as immune responses (Table 2).
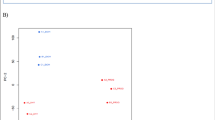
Transcriptome correlation and Esrrb altered mRNAs. a Transcriptome correlation analysis was performed using Spearman Ranking Correlation. Color represents the correlation coefficient. DY131 treatment to DU145-Esrrb cells results in the lowest correlation coefficient with DU145-pc3.1 cells. b Dot plot of Esrrb-induced gene expression alteration. Genes expressed at adequate level are tested for differential gene expression test. The plot was made by plotting the Log2FC (fold change) against the Log2 cpm (count-per-million) difference. Red color marks the genes that are significant differentially expressed (FDR < 0.05), and the blue lines marked the Log2FC cutoff value (Log2FC > 1 or Log2FC < −1). 67 genes passed both thresholds
Esrrb-regulated mRNA validation. Left panel qPCR validation of Esrrb-regulated mRNAs. Expression values were firstly normalized to Gapdh and normalized ratios are further normalized to that of DU145-pc3.1. Error bars represent standard deviation. Student t test was performed for statistical analysis (*p < 0.05). Seven genes were differentially expressed in both RNA-seq and qPCR, 1 gene, TGFbeta, is not differentially expressed in either assay and serves as a negative control. Right panel RNA-Seq analysis result, fold change (FC) indicates the ratio of normalized read counts in DU145-Esrrb to that of DU145-pc3.1
DY131 requires Esrrb to affect gene expression
To get a more comprehensive understanding of Esrrb-regulated genes and characterize Esrrb’s potential ligand dependent activity, control DU145-pc3.1 and DU145-Esrrb cells were treated with Esrrb/Esrrg synthetic ligand DY131. Since both qPCR and RNA-seq show Esrrb transcript concentration is extremely low in DU145 cells and Esrrg is absent, and Esrrb protein concentration is also below the detection limit of western-blot analysis, it was not surprising to observe DY131 treatment without Esrrb expressed did not result in any gene differentially expressed (Fig. 4a). After we applied DY131 to DU145-Esrrb cells, we found DY131 treatment most significantly modified the transcriptome (Figs. 2a, 4b). Further comparison of DU145-Esrrb cells alone to DY131-treated DU145-Esrrb cells detected 1161 altered mRNAs (861 down-regulated, 300 up-regulated). 15 of them overlapped with Esrrb-induced mRNA alterations (Fig. 4c, d; Table 3). We defined an Esrrb agonist as a ligand that moves the mRNA concentration in the direction as Esrrb does; and an antagonist moves the concentration in the opposite direction as Esrrb does. By comparing the trend of the altered genes induced by Esrrb expression and DY131 treatment, DY131 acts as an agonist for 4 of the 15 genes, and an antagonist for 11 of the 15 genes (Fig. 4d). There are another 1146 mRNAs changed with both Esrrb and DY131 treatment compared to Esrrb alone, indicating their responses is ligand-dependent (Table 3).
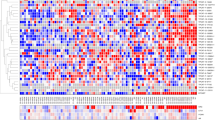
mRNA alteration by DY131 requires Esrrb expression. a DY131 treatment alone did not alter the expression of any gene. In contrast, when Esrrb was expressed, b DY131 altered 1161 mRNAs. c Venn Diagram of pairwise comparisons of altered mRNAs showed 15 (p = 0.0014) Esrrb altered mRNAs can be further regulated by DY131 treatment (overlap between Esrrb vs. control and Esrrb + DY131 vs. Esrrb). d Heat map of mRNA concentration of the 15 genes that response to both Esrrb expression as well as DY131 treatment. Log2-transformed normalized read counts of these 15 genes were color coded. DY131 is an agonist for 4 mRNAs that are responsive to Esrrb, while it is an antagonist of Esrrb in regulating the other 11 mRNAs
GO analysis showed Esrrb-dependent DY131 up-regulated genes were important for regulation of transcription, regulation of apoptosis and proliferation, and a majority of down-regulated genes are related to oxidation and reduction, metabolism and translation elongation (Table 4; Additional file 1: Table S1).
Discussion
Esrrb has gained lots of attention in recent years because of its biological function in stem cells and its ability to reprogram somatic cells to iPSC with oct4 and sox2 [6, 13, 17–21]. Several other functions of Esrrb have also been discovered including alteration of energy balance, estrogen receptor and glucocorticoid receptor transcription function modulation, Keap1-Nrf2 signaling inhibition, and tumorigenesis in prostate cancer and endometrial adenocarcinoma [9–12, 22–25]. But transcriptome-wide Esrrb function and Esrrb-regulated genes in cancer cells are not well studied.
Esrrb was reported by Chan et al. as a tumor suppressor in DU145 and LNCaP prostate cancer cells using both in vitro and in vivo models [9]. Expression of Esrrb induced p21/cdkn1a by directly binding to an ERRE in p21/cdkn1a’s promoter, arrested cell cycle at S-phase, and significantly inhibited cell growth [9, 26]. Interestingly, we did not find p21/cdkn1a up-regulation after Esrrb expression alone, but after we treated DU145 cells with 3 μM DY131, we observed a significant increase of p21/cdkn1a mRNA (Table 3; Additional file 2: Figure S1). Scrutinizing the data revealed that Chan’s lab cultured their cells with full serum, while we used charcoal-stripped serum for cell culture and DY131 treatment [9]. This implies that there is a compound or factor that can be removed by charcoal treatment modulated Esrrb’s activity [27, 28].
From the Esrrb-regulated gene list, we found a few target genes that are related to the known function of Esrrb. Kiaa1199 encoded gene product has been shown to associate with cellular mortality. A kiaa1199 mutation was reported to relate to nonsyndromic hearing loss. Considering the significant effect of Esrrb mutations on human hearing loss, kiaa1199 could be a mediator of Esrrb mutant related hearing loss [29–32] [33–36]. Another interesting Esrrb responsive gene is tagln (Transgelin). It was inhibited by Esrrb while DY131 treatment relieved the inhibition. Tagln was reported to promote DU145 cell migration and invasion, indicating Esrrb can also affect DU145 cell behavior by affecting tagln [37].
Judging by the numbers of altered genes induced by Esrrb with or without DY131, and the result that DY131 did not alter any mRNA in the absence of Esrrb, we conclude that DY131 activity is Esrrb-dependent.
Conclusions
In conclusion, we characterized the transcriptome alteration induced by Esrrb expression as well as Esrrb with its ligand DY131 in prostate cancer cells. We conclude Esrrb-target synthetic ligand requires Esrrb to generate its gene expression modulation effect. Finally, analysis of Esrrb target genes indicates Esrrb may be an important factor in regulating cell proliferation.
Methods
Cell culture and reagents
DU145 (ATCC Number: HTB-81) and HEK293 (ATCC number: CRL-1571) cells were obtained from the American Type Culture Collection (ATCC). DU145 cells were cultured in RPMI1640 media (Invitrogen, Grand Island, NY, USA) with 10 % Fetal Bovine Serum (FBS) (GE Healthcare Life Sciences, Logan, UT, USA). HEK293 cells were cultured in Eagle’s Minimal Essential Medium (DMEM) (Invitrogen, Grand Island, NY, USA) with 10 % FBS. 70 % confluent DU145 cells were transfected with either pcDNA3.1-zeo (+)-Esrrb expression vector [4], or control empty vector pcDNA3.1-zeo (+) (Promega, Madison, WI, USA). Empty vector or Esrrb expression vector transfected DU145 cells were maintained in medium containing 150 μg/ml Zeocine (Invitrogen, Grand Island, NY, USA) for 3 weeks for selection. Two biological replicates of DU145 cells transfected with Esrrb were pooled together respectively and were named DU145-Esrrb. Two biological replicates of DU145 cells transfected with control vector were pooled together respectively and were named DU145-pc3.1. Total RNA and protein were collected from cells after they are confluent in 60 mm petri dishes, cultured with phenol-red free RPMI1640 with 10 % Charcoal-stripped FBS [38]. For DY131 (Tocris Bioscience, Bristol, UK) treatment, cells are plated in 60 mm petri dishes until confluent; DU145-pc3.1 and DU145-Esrrb are incubated with 3 μM DY131 diluted in medium with charcoal-stripped FBS for indicated length of time.
Western-blot
Total protein was isolated from DU145-pc3.1, and DU145-Esrrb cells. 20 μg protein was loaded on 9 % SDS gels. After the proteins were transferred to nitrocellulose membrane, the membrane was blocked and then incubated with 1:2000 diluted monoclonal anti-Esrrb mouse IgG (R&D system, Cat. no: PP-H6705-00) and 1:2000 diluted polyclonal anti-GPADH rabbit IgG (Santa Cruz, Dallas, TX, USA, Cat. no: sc-25777) at 4 degrees overnight. The membrane was then washed and incubated with anti-mouse or anti-rabbit secondary antibody. Chemoluminescence (Promega, Madison, WI, USA) signals were collected using x-ray films (Fisher Scientific, Pittsburg, PA, USA).
Reverse transcriptase PCR and quantitative PCR
Total RNA was isolated and purified from DU145-pc3.1 and DU145-Esrrb using RNeasy kit (Qiagen, Venlo, Netherlands). 1000 ng of total RNA was used to create cDNA libraries using Superscript III Reverse Transciptase with random primers and oligodT (Invitrogen, Grand Island, NY, USA). Esrrb mRNA concentration was determined using quantitative PCR (qPCR) (iQ SYBR, BioRad, Hercules, CA, USA) on ABI7500 system (Applied Biosystems, Foster City, CA, USA). PCR condition: 95°, 30 s; 60°, 40 s; 72°, 40 s. Each qPCR test was performed three times on each of the two biological replicates. Primer sequences: zcwpw2 (Genbank: NM_001040432): forward primer: AACAGGGTTGTCTGTGAGACGGA; reverse primer: TGCAGGAGCTTCTGGGCTGC. hoxb8 (Genbank: NM_024016): forward primer: GATGCGCC CGCAAGCAGC; reverse primer: CCCAGGGCGTGCGATACCTC. tagln (Genbank: NM_001001522): forward primer: ATGCCCCGGATGACTTGGCT; reverse primer: GCCATGTCTGGGGAAAGCTCCT. f13a1 (Genbank: NM_000129): forward primer: TGTTCCGTGAAATCCGGCCC; reverse primer: TGCACGTCCAG CTCGCCATA. pxdn (Genbank: NM_012293): forward primer: GCAAGCATTTAA GGGACTTGCCTCT; reverse primer: GCAAAAATAGCCTCTCGAGCTTCGG. aox1 (Genbank: NM_001159): forward primer: TACGTGAACGGCCGCAAGGT; reverse primer: TGGCTGGGTGATGCCTTATCCT. bmp4 (Genbank: NM_001202): forward primer: CCACCACGAAGAACA TCTGGAG; reverse primer: GCCCCTTTCCCAATCAGGGC. tgfβ: (Genbank: NM_000660) forward primer: AGTGGACATC AACGGGTTCAC; reverse primer: CGCACGCAGCAGTTCTTCTC. gapdh: (Genbank: NM_001256799); forward primer: ACCCACTCCTCCACCTTTG; reverse primer: CTCTTGTGCTCTTGCTGGG. Esrrb: (Genbank: NM_004452) forward primer: CAAGAAGCTCAAGGTGGAGAAGGAGGAG; reverse primer: CGGTCTGTCC GTTTGTCTGTCTGTAGGT. Esrrg: (Genbank: NM_001134285) forward primer: ACCATGAATGGCCATCAGA A; reverse primer: ACCAGCTGAGGGTTCAGGTAT.
Deep sequencing and differentially expressed genes
2500 ng total RNA from two biological replicates was used to generate cDNA libraries using TruSeq Stranded mRNA Sample Preparation kits (Illumina, San Diego, CA, USA). RNA quality and fragment sizing of cDNA library were determined by the University of Missouri DNA core. Deep sequencing was performed by the MU DNA core using Illumina HiSeq 2000 following the manufacture’s instruction. Briefly, samples (8 total) were pooled into one lane with each sample annealed to a specific indexed adaptor. 50 bp single end reads were generated. For each sample, approximately 18 million reads were generated in.fastq format (NCBI-GEO, accession number: GES71208). The sequencing reads were trimmed and filtered using FASTX-Toolkit (V 0.0.13) (http://hannonlab.cshl.edu/fastx_toolkit), and mapped to genome (UCSC hg18) using TopHat2 [39, 40]. Gene expression values were determined by gene raw read counts using an in-house tool MULTICOM-MAP [41–43]. Raw reads were normalized to each sample’s library size and differentially expressed genes were calculated using R/Bioconductor package edgeR [44]. Specifically, we kept the genes that have at least 1 count-per-million (cpm) in at least 2 samples and computed the effective library sizes. Pairwise gene expression tests were carried out using exact test. Differentially expressed genes were determined by log2 fold change (Log2FC) (Log2FC ≥ 1, or Log2FC ≤ −1), p value (p < 0.05) and false discovery rate (FDR < 0.05) [45].
Gene set function enrichment
Gene ontology (GO) analysis was performed using DAVID bioinformatics sources 6.7 [46, 47]. Differentially expressed genes from certain pairwise comparisons were uploaded to DAVID server (http://david.abcc.ncifcrf.gov) and GO analysis were performed for biological process (BP). Minimum counts were set as default value (two counts) and maximum EASE score (p value) was set to 0.05. Differentially expressed genes pathway enrichment analysis was performed using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway [48, 49]. Gene expression profiles Spearman ranking correlation analysis was analyzed using R (version 3.0.2). Gene expression heat map and hierarchical clustering were created by R/Bioconductor (version 2.13) package gplot.
Statistical analysis
qPCR experiments were performed in triplicate on both biological replicates. T test was employed to statistically analyze whether the differences in gene expression is significant (p < 0.05). Statistical significance of gene set overlap (Venn Diagram) are tested according to previous reported method [17].
Availability of supporting data
The data sets supporting the results of this article are available in the NCBI-GEO repository, accession number: GSE71208, URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE71208.
Abbreviations
- Esrrb:
-
estrogen related receptor beta
- ERRE:
-
estrogen related receptor response element
- SFRE:
-
steroid factor response element
- iPSC:
-
induced pluripotent stem cells
- OSKM:
-
Oct4, Sox2, Klf4, cMyc
- FBS:
-
fetal bovine serum
- RT:
-
reverse transcriptase
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- GO:
-
gene ontology
- FC:
-
fold change
- Esrrg:
-
estrogen related receptor gamma
References
Giguere V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–4.
Vanacker JM, Pettersson K, Gustafsson JA, Laudet V. Transcriptional targets shared by estrogen receptor-related receptors (ERRs) and estrogen receptor (ER) alpha, but not by ERbeta. EMBO J. 1999;18:4270–9.
Luo J, Sladek R, Bader JA, Matthyssen A, Rossant J, et al. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-beta. Nature. 1997;388:778–82.
Zhou W, Liu Z, Wu J, Liu JH, Hyder SM, et al. Identification and characterization of two novel splicing isoforms of human estrogen-related receptor beta. J Clin Endocrinol Metab. 2006;91:569–79.
Welstead GG, Brambrink T, Jaenisch R. Generating iPS cells from MEFS through forced expression of Sox-2, Oct-4, c-Myc, and Klf4. J Vis Exp. 2008;14.
Feng B, Jiang J, Kraus P, Ng JH, Heng JC, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11:197–203.
Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–22.
Cheung CP, Yu S, Wong KB, Chan LW, Lai FM, et al. Expression and functional study of estrogen receptor-related receptors in human prostatic cells and tissues. J Clin Endocrinol Metab. 2005;90:1830–44.
Yu S, Wong YC, Wang XH, Ling MT, Ng CF, et al. Orphan nuclear receptor estrogen-related receptor-beta suppresses in vitro and in vivo growth of prostate cancer cells via p21(WAF1/CIP1) induction and as a potential therapeutic target in prostate cancer. Oncogene. 2008;27:3313–28.
Fujimura T, Takahashi S, Urano T, Ijichi N, Ikeda K, et al. Differential expression of estrogen-related receptors beta and gamma (ERRbeta and ERRgamma) and their clinical significance in human prostate cancer. Cancer Sci. 2010;101:646–51.
Zhou W, Lo SC, Liu JH, Hannink M, Lubahn DB. ERRbeta: a potent inhibitor of Nrf2 transcriptional activity. Mol Cell Endocrinol. 2007;278:52–62.
Bombail V, Collins F, Brown P, Saunders PT. Modulation of ER alpha transcriptional activity by the orphan nuclear receptor ERR beta and evidence for differential effects of long- and short-form splice variants. Mol Cell Endocrinol. 2010;314:53–61.
Chen X, Xu H, Yuan P, Fang F, Huss M, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17.
Ouyang Z, Zhou Q, Wong WH. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc Natl Acad Sci USA. 2009;106:21521–6.
Percharde M, Lavial F, Ng JH, Kumar V, Tomaz RA, et al. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes Dev. 2012;26:2286–98.
Nishiyama A, Sharov AA, Piao Y, Amano M, Amano T, et al. Systematic repression of transcription factors reveals limited patterns of gene expression changes in ES cells. Sci Rep. 2013;3:1390.
Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:16438–43.
Carter MG, Stagg CA, Falco G, Yoshikawa T, Bassey UC, et al. An in situ hybridization-based screen for heterogeneously expressed genes in mouse ES cells. Gene Expr Patterns. 2008;8:181–98.
van den Berg DL, Zhang W, Yates A, Engelen E, Takacs K, et al. Estrogen-related receptor beta interacts with Oct4 to positively regulate Nanog gene expression. Mol Cell Biol. 2008;28:5986–95.
Zhang X, Zhang J, Wang T, Esteban MA, Pei D. Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells. J Biol Chem. 2008;283:35825–33.
Storm MP, Kumpfmueller B, Thompson B, Kolde R, Vilo J, et al. Characterization of the phosphoinositide 3-kinase-dependent transcriptome in murine embryonic stem cells: identification of novel regulators of pluripotency. Stem Cells. 2009;27:764–75.
Gao M, Sun P, Wang J, Zhao D, Wei L. Expression of estrogen receptor-related receptor isoforms and clinical significance in endometrial adenocarcinoma. Int J Gynecol Cancer. 2006;16:827–33.
Wang SC, Myers S, Dooms C, Capon R, Muscat GE. An ERRbeta/gamma agonist modulates GRalpha expression, and glucocorticoid responsive gene expression in skeletal muscle cells. Mol Cell Endocrinol. 2010;315:146–52.
Byerly MS, Al Salayta M, Swanson RD, Kwon K, Peterson JM, et al. Estrogen-related receptor beta deletion modulates whole-body energy balance via estrogen-related receptor gamma and attenuates neuropeptide Y gene expression. Eur J Neurosci. 2013;37:1033–47.
Byerly MS, Swanson RD, Wong GW, Blackshaw S. Estrogen-related receptor beta deficiency alters body composition and response to restraint stress. BMC Physiol. 2013;13:10.
Castet A, Herledan A, Bonnet S, Jalaguier S, Vanacker JM, et al. Receptor-interacting protein 140 differentially regulates estrogen receptor-related receptor transactivation depending on target genes. Mol Endocrinol. 2006;20:1035–47.
Sumi D, Ignarro LJ. Estrogen-related receptor alpha 1 up-regulates endothelial nitric oxide synthase expression. Proc Natl Acad Sci USA. 2003;100:14451–6.
Vanacker JM, Bonnelye E, Chopin-Delannoy S, Delmarre C, Cavailles V, et al. Transcriptional activities of the orphan nuclear receptor ERR alpha (estrogen receptor-related receptor-alpha). Mol Endocrinol. 1999;13:764–73.
Abe S, Usami S, Nakamura Y. Mutations in the gene encoding KIAA1199 protein, an inner-ear protein expressed in Deiters’ cells and the fibrocytes, as the cause of nonsyndromic hearing loss. J Hum Genet. 2003;48:564–70.
He QY, Liu XH, Li Q, Studholme DJ, Li XW, et al. G8: a novel domain associated with polycystic kidney disease and non-syndromic hearing loss. Bioinformatics. 2006;22:2189–91.
Guo J, Cheng H, Zhao S, Yu L. GG: a domain involved in phage LTF apparatus and implicated in human MEB and non-syndromic hearing loss diseases. FEBS Lett. 2006;580:581–4.
Michishita E, Garces G, Barrett JC, Horikawa I. Upregulation of the KIAA1199 gene is associated with cellular mortality. Cancer Lett. 2006;239:71–7.
Collin RW, Kalay E, Tariq M, Peters T, van der Zwaag B, et al. Mutations of ESRRB encoding estrogen-related receptor beta cause autosomal-recessive nonsyndromic hearing impairment DFNB35. Am J Hum Genet. 2008;82:125–38.
Ben Said M, Ayedi L, Mnejja M, Hakim B, Khalfallah A, et al. A novel missense mutation in the ESRRB gene causes DFNB35 hearing loss in a Tunisian family. Eur J Med Genet. 2011;54:e535–41.
Lee K, Khan S, Ansar M, Santos-Cortez RL, Ahmad W, et al. A novel ESRRB deletion is a rare cause of autosomal recessive nonsyndromic hearing impairment among Pakistani families. Genet Res Int. 2011;2011:368915.
Safka Brozkova D, Lastuvkova J, Machalova E, Lisonova J, Trkova M, et al. DFNB35 due to a novel mutation in the ESRRB gene in a Czech consanguineous family. Int J Pediatr Otorhinolaryngol. 2012;76:1681–4.
Lee EK, Han GY, Park HW, Song YJ, Kim CW. Transgelin promotes migration and invasion of cancer stem cells. J Proteom Res. 2010;9:5108–17.
Welshons WV, Grady LH, Engler KS, Judy BM. Control of proliferation of MCF-7 breast cancer cells in a commercial preparation of charcoal-stripped adult bovine serum. Breast Cancer Res Treat. 1992;23:97–104.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25.
Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11.
Sun L, Fernandez HR, Donohue RC, Li J, Cheng J, et al. Male-specific lethal complex in Drosophila counteracts histone acetylation and does not mediate dosage compensation. Proc Natl Acad Sci USA. 2013;110:E808–17.
Sun L, Johnson AF, Donohue RC, Li J, Cheng J, et al. Dosage compensation and inverse effects in triple X metafemales of Drosophila. Proc Natl Acad Sci USA. 2013;110:7383–8.
Sun L, Johnson AF, Li J, Lambdin AS, Cheng J, et al. Differential effect of aneuploidy on the X chromosome and genes with sex-biased expression in Drosophila. Proc Natl Acad Sci USA. 2013;110:16514–9.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Li H, Lovci MT, Kwon YS, Rosenfeld MG, Fu XD, et al. Determination of tag density required for digital transcriptome analysis: application to an androgen-sensitive prostate cancer model. Proc Natl Acad Sci USA. 2008;105:20179–84.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Authors’ contributions
YL established the model cell line and performed RNA isolation, Esrrb expression status test, western blot, cell growth assay, differentially expressed gene analysis, functional enrichment of gene sets by GO and KEGG pathway, construct gene regulation network and statistical analysis. YL, JL, JC performed sequence alignment and generated the gene expression count table for RNA-seq data analysis. YL and DBL conceived of the study, participated in its design and drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Wei Zhou for cloning Esrrb. We also thank Nicholas Starkey, Benjamin Merideth and Yufei Li for helpful discussions. This publication or project was made possible in part by Grant Number P50AT006273 from the National Center for Complementary and Integrative Health (NCCIH), the Office of Dietary Supplements (ODS), and the National Cancer Institute (NCI). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCCAM, ODS, NCI, or the National Institutes of Health.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional files
12867_2015_49_MOESM2_ESM.pdf
Additional file 2: Table S1. Gene ontology analysis result. Table S2. Esrrb expression with DY131 treatment (control vs. Esrrb + DY131).
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lu, Y., Li, J., Cheng, J. et al. Messenger RNA profile analysis deciphers new Esrrb responsive genes in prostate cancer cells. BMC Molecular Biol 16, 21 (2015). https://doi.org/10.1186/s12867-015-0049-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12867-015-0049-1