Abstract
Background
Macrobrachium rosenbergii, is one of a major freshwater prawn species cultured in Southeast Asia. White tail disease (WTD), caused by Macrobrachium rosenbergii nodavirus (MrNV), is a serious problem in farm cultivation and is responsible for up to 100% mortality in the post larvae stage. Molecular data on how M. rosenbergii post-larvae launches an immune response to an infection with MrNV is not currently available. We therefore compared the whole transcriptomic sequence of M. rosenbergii post-larvae before and after MrNV infection.
Results
Transcriptome for M. rosenbergii post-larvae demonstrated high completeness (BUSCO Complete: 83.4%, fragmentation: 13%, missing:3.3%, duplication:16.2%; highest ExN50 value: 94%). The assembled transcriptome consists of 96,362 unigenes with N50 of 1308 bp. The assembled transcriptome was successfully annotated against the NCBI non-redundant arthropod database (33.75%), UniProt database (26.73%), Gene Ontology (GO) (18.98%), Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups (EggNOG) (20.88%), and Kyoto Encyclopedia of Genes and Genome pathway (KEGG) (20.46%). GO annotations included immune system process, signaling, response to stimulus, and antioxidant activity. Differential abundance analysis using EdgeR showed 2413 significantly up-regulated genes and 3125 significantly down-regulated genes during the infection of MrNV.
Conclusions
This study reported a highly complete transcriptome from the post-larvae stage of giant river prawn, M. rosenbergii. Differential abundant transcripts during MrNV infection were identified and validated by qPCR, many of these differentially abundant transcripts as key players in antiviral immunity. These include known members of the innate immune response with the largest expression change occurring in the M. rosenbergii post-larvae after MrNV infection such as antiviral protein, C-type lectin, prophenol oxidase, caspase, ADP ribosylation factors, and dicer.
Similar content being viewed by others
Background
Macrobrachium rosenbergii, known as the giant freshwater prawn, is an economically important crustacean species in Southeast Asia. Production of M. rosenbergii increased dramatically over the last 20 years and has exceeded 200,000 t per year since 2002 [1]. A major problem in the cultivation of M. rosenbergii relates to loss through infectious diseases caused by viruses, bacteria, and fungi [2]. One of the most serious viral threats to M. rosenbergii culture is Macrobrachium rosenbergii nodavirus (MrNV) that causes white tail disease (WTD). This disease was first discovered in Guadeloupe Island [3] and subsequently in Taiwan [4], China [5], India [6], Thailand [7], and Australia [8]. MrNV is a non-enveloped, icosahedron with 26–27 nm diameter composed of a nucleocapsid bearing two positive single-stranded RNA genomes (RNA-1 and RNA-2). RNA-1 is 3202 bp - ssRNA encoding protein A or RNA-dependent RNA polymerase (RdRp) and protein B2 [9]. Protein B2 is capable of inhibiting the RNAi pathway of the host cell [10]. RNA-2 is 1175 bp - ssRNA encoding capsid protein [11]. The prominent sign of infected post larvae (PL) is the appearance of whitish muscle commonly in abdominal region. Mortalities may reach 100% within 7–15 days after the infection or 3–5 days after the appearance of the first anatomical signs [6]. However, experimental transmission of MrNV revealed that the virus failed to cause mortality in adult prawns [6]. As there is no cure for WTD infected prawns, preventive procedures have been implemented such as screening of brood stock and PL, and good farm management [7, 12, 13]. Understanding the prawn’s immune response specifically to WTD may also provide bio-rationale targets to help contain and restrict disease outbreak.
As an arthropod crustacean, the innate immune system of M. rosenbergii is composed of both humoral and cellular responses. Humoral immune responses include up-regulation of the prophenol oxidase system (ProPO), clotting proteins, melanization and antimicrobial peptides [14]. Cellular immune responses involve hemocyte activities such as phagocytosis, apoptosis, nodule formation, and encapsulation which function to neutralize pathogens [15]. Indirect recognition of pathogens or pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) such as Toll receptors leads to the activation of these humoral and cellular immune responses [16].
In recent years, high-throughput technology such as next generation sequencing (NGS) has emerged and is widely used in both genomic and transcriptomic research. NGS technology can also be used to study differential gene-expression on various tissues or certain conditions such as stress and pathogen infection [17] and in both model, and non-model organisms [18]. There are transcriptomic studies for penaeid shrimps such as Peneaus monodon [19,20,21], Litopeneaus vannamei [22,23,24], Fenneropeneaus chinensis [25, 26], Fenneropenaeus merguiensis [27, 28], and Marsupeneaus japonicus [29] to investigate tissue-specific expression, the stress response, and viral infection. Moreover, many studies have been performed on whole transcriptome sequencing of the hepatopancreas of M. rosenbergii in response to Vibrio parahaemolyticus infection [30], hepatopancreas and lymphoid organ in response to white spot syndrome virus (WSSV) [31, 32], and intestinal tissue in response to WSSV or the viral PAMP mimic (poly I:C) [33]. Most recently, transcriptomic analysis of hematopoietic tissue of M. rosenbergii adult prawn in response to MrNV infection has been studied [34]. Many of differentially abundant transcripts were belonged to various immune mechanisms such as pattern-recognition receptors, antioxidants, and antimicrobial peptides [34].
MrNV has direct impact on M. rosenbergii post-larvae culture. However, there is no transcriptomic data on M. rosenbergii post-larvae in response to the infection with MrNV. To address this deficiency, the whole transcriptome from six biological replicates of each treatment from healthy and MrNV-infected PL were sequenced. In this study, a highly complete transcriptome for M. rosenbergii post-larvae was assembled and annotated. This transcriptome can be used as reference transcriptome for the further gene expression analysis of M. rosenbergii. Differential abundant transcripts were examined and immune-related genes in response to MrNV infection were reported. In addition, differential abundance and RNAseq results were validated by quantitative PCR in separate biological samples.
Results
Sequence read data and raw data pre-processing
RNA sequencing for two groups with six biological replicates (n = 12) produced a total of 522,296,142 raw reads with an average of 43.5 ± 5.1 (mean ± SD) million read pairs per sample (Table 1). The raw reads were subjected to quality trimming which included adaptor removal. After quality trimming, a total of 501,900,423 (96.09%) 65 bp trimmed reads had high average Phred score (96.13% with Phred ≥30).
De novo transcriptome assembly and quality assessment
The trimmed reads were subjected to de novo transcriptome assembly using Trinity. The trimmed reads were further reduced into 51,971,920 reads (10.36%) during in silico normalization prior to de novo assembly. The Trinity assembler produced 109,616 which were then clustered into 96,362 unigenes with N50 of 1308 bp, and mean length of 776.73 bp by CD-HIT software. The transcriptome had high overall fragment mapping rate of 96.85% with 90.08% that aligned concordantly ≥1 times. Based on BUSCO, the assembled transcriptome was highly complete with 889 (83.4%) ortholog gene from Arthropoda database with low fragmented and missing BUSCOs (C:83.4%[S:67.2%,D:16.2%],F:13.3%, M:3.3%,n:1066). In addition, the ExN50 statistics showed that the maximum N50 value was on E94% with 2031 bp N50 length. The summary of the transcriptome assembly and quality assessment are shown in Table 2.
Functional annotation
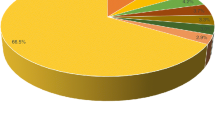
The assembled transcripts were subjected to the homology search by Blastx software using UniProt and non-redundant arthropod database. Blastx search against UniProt database yielded 25,761 (26.73%) significant hits (E-value < 1-e5). The majority of top-hit species distribution were Homo sapiens with 5587 (21.7%) hits followed by Mus musculus and Drosophila melanogaster with 4234 (16.4%) and 4043 (15.7%) hits, respectively (Fig. 1a).
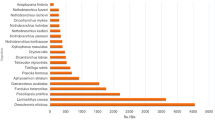
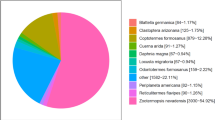
In case of non-redundant arthropod database, Blastx search yielded 32,523 (33.75%) significant hits (E-value < 1-e5). Top-hit species distribution was mainly dominated by Hyalella azteca with 11,197 (34.4%) hits followed by Cryptotermes secundus with 1841 (5.6%) hits (Fig. 1b). Prawn species including Litopenaeus vannamei, M. nipponense, M. rosenbergii, Penaeus monodon, and M. japonicus were the eighth, eleventh, thirteenth, seventeenth, and eighteenth top-hits, respectively, as demonstrated in top-hit crustaceans species distribution (Fig. 1c). Annotation results of assembled transcriptome are available in Additional file 1: Tables S1 and S2.
Gene ontology mapping
Functional annotations including EggNOG (Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups), KEGG (Kyoto Encyclopedia of Genes and Genomes), and GO (Gene Ontology) were obtained from Blastx UniProt results using Trinotate suite. Total of 18,291 unigenes were GO mapped. Total of 187,582 GO assignments (level 2) were generated from GO annotations and divided into three GO domains including cellular components (75,535 or 40.27%), molecular functions (25,373 or 13.51%), and biological processes (86,674 or 46.20%) (Fig. 2). Among the cellular component domains, annotated unigenes were mostly involved in “cell” (14,523) and “cell part” (14,499) followed by “organelle” (11,625). The least abundance in cellular component were “synapse part” (505) followed by “supramolecular complex” (626) (Fig. 2). Molecular function domains were primarily dominated by “binding” (12,177) and “catalytic activity” (7378). The least abundance in molecular function were “molecular carrier activity” [35] followed by “antioxidant activity” [36] (Fig. 2). In the biological process domains, annotated unigenes were mostly involved in “cellular process” (13,234) followed by “metabolic process” (10,855). The least abundance in biological process were “behavior” (683) followed by “immune system process” (1219) (Fig. 2).
EggNOG classification showed that 20,130 unigenes were identified in the EggNOG database. Total of 20,704 EggNOG functional annotations were obtained and classified into 23 categories (Fig. 3). The most abundant category was “Function unknown” (9054 or 43.73%) The second most abundant was “Post-translational modification, protein turnover, chaperones” (2000 or 9.66%) followed by “Intracellular trafficking, secretion, and vesicular transport” (1614 or 7.79%), and “Signal transduction mechanisms” (1302 or 6.29%), respectively. The least abundant category was “Nuclear structure” (2 or 0.01%) followed by “Cell motility” (4 or 0.02%) (Fig. 3).
A total of 19,715 unigenes were matched to the KEGG database. Of those, 7917 unigenes had orthologs in the KEGG orthology database. A total of 14,289 KEGG orthology (KO) were obtained from those unigenes and then categorized into five major categories including “Metabolism” (3171 or 22.19%), “Genetic information processing” (2351 or 16.45%), “Environmental information processing” (3203 or 22.42%), “Cellular processes” (1522 or 10.65%), and “Organismal systems” (4042 or 28.29%) (Fig. 4). KO distribution results showed that the most abundance orthology was signal transduction from “Environmental information processing” category with 1779 unigenes. The second most abundance was translation from “Genetic information processing” category (1078 unigenes) followed by transport and catabolism from “Environmental information processing” category (1055 unigenes), respectively (Fig. 4).
Differential abundance analysis
To identify differentially abundant transcripts between two groups, transcripts that had count per millions (CPM) more than 1 in at least two samples were selected before the analysis. Total of 31,377 transcripts survived the cut-off and were subjected to TMM normalization. The differential abundance analysis using EdgeR was performed followed by Benjamini-Hochberg method for multiple p-value correction. Total of 5538 transcripts were reported to be differentially expressed (FDR < 0.05, LogFC < ±1). Of those, 2413 transcripts were significantly up-regulated and 3125 transcripts were significantly down-regulated after the infection of MrNV. Full-list of differentially abundant transcripts is available in Additional file 2: Table S3. Summary of the transcriptome assembly, annotation and differential abundance analysis were listed in Table 3.
Among those differentially abundant transcripts, various transcripts were reported to be involved in immune system in response to viral infection (Table 4). These transcripts were categorized into 13 functional groups including antiviral protein (1 unigene), antimicrobial protein (9 unigenes), pattern recognition proteins (19 unigenes), toll-signaling pathway (3 unigenes), RNAi pathway (2 unigenes), prophenol oxidase system (4 unigenes), serine proteinase cascade (5 unigenes), ubiquitin proteasome pathway (4 unigenes), antioxidant system (5 unigenes), blood coagulation (2 unigenes), apoptosis (3 unigenes), phagocytosis (7 unigenes), and other immune genes (10 unigenes) as listed in Table 4. From the list, total of 56 unigenes were reported to be significantly up-regulated, whereas 18 unigenes were significantly down-regulated.
To examine the homogeneity across biological replicates, principle component analysis (PCA) was performed. Transcripts with extremely low abundance (sum of read count < 10) were filtered out. Total of 84,092 transcripts survived the cut-off and were subjected to the PCA. The PCA results showed strong clustering within each group. The both groups formed distinct clusters within principle component 1 (PC1) which responsible for 42.62% of the variance in the expression (Fig. 5). In addition, the top 100 most differentially abundant transcripts were clustered using Pearson’s correlation and displayed in heatmap (Fig. 6). The biological replicates were clustered within the same group and demonstrate clear distinction between control and infected group.
Quantitative RT-PCR
To validate differential abundance results from the RNAseq pipeline, qRT-PCR was performed using nine selected genes from the list of DEG involved in the immune system. Elongation factor1-alpha (EF1-alpha) was used as an internal reference gene. Four separate biological replicates from each group were used in two-step qRT-PCR. The abundance levels were calculated using the delta-delta Ct method. The qRT-PCR results showed that all up-regulated genes had greater differences in transcript abundance than those of RNAseq, whereas all down-regulated genes had smaller differences (Table 5). According to the qRT-PCR results, all of the selected genes were differentially abundant after the infection of MrNV (p > 0.05) except Spz which had p-value of 0.053 (Table 5). Comparative heatmaps were generated using relative abundance (qRT-PCR) and transcript per millions (TPM) for RNAseq results (Fig. 7a). Expression patterns of all selected genes were comparable between qRT-PCR and RNAseq. Furthermore, the Pearson’s correlation coefficients were calculated using the average log-fold change ratio between the two methods and demonstrated highly significant correlation as shown in Fig. 7b (R2 = 0.9531).
Comparison of fold change in gene expression using either qRT-PCR or RNAseq. a Heatmap representing transcript per million (TPM) expression from RNAseq and relative expression from qRT-PCR. The Z-score scales are shown in the top-right corner ranging from blue to yellow. b Regression plot demonstrating the direct correlation between the average log2 FC expression values from both RNAseq and qRT-PCR. c The qRT-PCR validation results of nine selected genes including anti-lipopolysaccharide factor 1 (ALF1), Spatzle (Spz), copper/zinc superoxide dismutase 3 (CuZnSOD3), caspase (CASP), antiviral protein (Anv), dicer (DICER), hemicentin-1-like (HMCN1), ADP ribosylation factors (ARF), and prophenoloxidase (ProPO) with elongation factor1-alpha (EF1-alpha) as an internal reference
Discussion
In this study, we report a highly complete transcriptome for M. rosenbergii post-larvae and identified immune-related genes in response to MrNV infection using NGS platform as well as verified the transcript abundance of selected genes using qRT-PCR. We not only present the transcriptome data, but we have published our computational pipeline to benefit the wider scientific community. Raw data has been uploaded to the National Centre for Biotechnology Information Sequence Read Archive (SRA) under the accession BioProject number: PRJNA550272. The data analysis pipeline is available on GitHub at https://github.com/prawnseq/Mrosenbergii_MrNV_RNAseq.
The transcriptome of giant fresh water prawn (M. rosenbergii) was assembled to expand the transcriptomic resources for further gene expression analysis of this species. Therefore, we aimed for maximizing transcriptome coverage while minimizing the redundancy to generate high quality transcriptome. The BUSCO results showed that the assembled transcriptome was highly complete (C:83.4%) with low fragmentation (F:13%), missing (M:3.3%), and duplication (D:16.2%). These results were comparable to the assembled transcriptome from L. vannamei (C:98.0%, F:0.7%, M:1.3%, D:25.5%) [37] and from P. monodon (C:98.2%, F:0.8%, M:1.0%, D:51.3%) [20]. According to Blastx results against UniProt database, the top hits were mammalian species (H. sapiens, M. musculus) followed by D. melanogaster because UniProt database is a comprehensive annotation database which annotated and reviewed by the experts [38]. Therefore, majority of UniProt subjects were obtained from well-studied model organisms such as human, mouse and fruit fly. However, according to Blastx results against Nr-Arthropod database, majority of the top hits were matched to crustacean species, Hyalella Azteca. Previous transcriptomic studies of M.rosenbergii performed de novo assembly separately from each sample and then clustered into a global transcriptome [30,31,32,33]. In this study, all read data were merged before the assembly, resulting in an increase in the abundance of low expressed transcripts, therefore increasing coverage of the assembled transcripts [39]. In addition, the highest ExN50 value at 94% indicated that the assembled transcripts had high coverage and was assembled from sufficient read data [40].
To identify differentially abundance transcripts associated with MrNV infection, we performed RNA sequencing on six replicates of each healthy PL and experimentally MrNV infected PL. The results showed that 5538 transcripts were differentially expressed with 2413 transcripts were up-regulated and 3125 transcripts were down-regulated. Among those transcripts, some of these were involved in the innate immune system in response to viral infection.
To validate our transcript assembly and differential abundance results, we also selected 9 targets for validation with qPCR, and demonstrated a very low false discovery rate in these genes. We filtered our list of DEGs according to functional groups of importance including pattern recognition proteins (PRPs) and antiviral protein, prophenol oxidase (ProPO) system, the Toll-IMD signaling pathways, antimicrobial peptides (AMPs) and blood clotting system, phagocytosis and apoptosis, antioxidant system, and RNA interference (RNAi). Numerous studies demonstrated high correlation between RNAseq and qPCR data [41,42,43]. Moreover, RNAseq studies of M. rosenbergii in response to bacterial and viral infection also showed high correlation between RNAseq and qPCR results [30,31,32]. To enhance our confidence, we validated these results in independent biological samples.
Crustacean innate immune system requires the recognition of the pathogens by pattern recognition proteins (PRPs) to trigger the immune responses [44]. PRPs are groups of germ-line encoded proteins that can activate humoral and cellular immune response via immune signaling pathway [45]. Several studies identified prawn PRPs and examined its role in prawn immune system regarding viral infection. M. rosenbergii C-type lectin (MrCTL) expression in hepatopancreas was up-regulated after a challenge with Vibrio parahaemolyticus or white spot syndrome virus (WSSV) [46]. Ficolin expression in M. rosenbergii hepatopancreas was found to be up-regulated after the infection of V. anguillarum and WSSV [47]. Moreover, M. rosenbergii mannose-binding lectin (MBL) expression in gills was also up-regulated in response to WSSV or MrNV infection [48]. In this study, various PRPs such as C-type lectin, ficolin, and antiviral protein which are part of C-type lectin family were differentially expressed after the infection of MrNV suggesting that these PRPs play important roles in immune system against MrNV infection. Importantly, we validated the expression of one of these genes, antiviral protein, which was up-regulated by 2.48-fold in the RNAseq and 8.97-fold by qPCR (Table 5). However, in our results, MBL expression was down-regulated which contradicts the previous report [48]. This maybe because MBL is not required for antiviral response of post-larvae prawn against MrNV or the expression of MBL in post-larvae prawn being suppressed by MrNV. Further investigation is needed to understand the effects of MrNV to MBL expression in post-larvae stage.
Prophenol oxidase (ProPO) activating system participates in first line of immune system by triggering melanization and other responses such as hemocyte induction, encapsulation, and nodule formation [14, 49, 50]. Recognition of PAMPs by PRPs leads to activation of serine proteinases cascade, resulting in the production of active PO enzyme. The active PO enzyme produces polymeric melanin around invading pathogens resulting in melanization [50]. The RNAseq presented here shows a 2.01-fold up-regulation of ProPO expression in response to MrNV infection in which the expression was validated by qPCR (9.38-fold up-regulation; Table 5). In addition, three types of prophenoloxidase activating enzyme were also up-regulated in RNAseq results. These results indicate involvement of ProPO-activating system in MrNV infection.
The Toll-IMD signaling pathways are considered to be the most crucial immune signaling pathways in invertebrates [51]. The toll receptor is triggered by cytokine-like ligand Spätzle whereas vertebrate Toll-like receptors (TLRs) recognize pathogens directly [52, 53]. Triggering the toll receptor leads to activation of NF-kappa-B family protein Dif/Dorsal and then leads to up-regulation of immune-related genes such as antimicrobial peptide (AMPs) [53]. Toll receptor from M. rosenbergii have been identified [54, 55] and was found to be gradually up-regulated in gills during the WSSV challenge [55]. M. rosenbergii spätzle protein has been found to be up-regulated in hemocytes after the bacteria infection [56]. In addition, spätzle protein in F. chinensis was up-regulated after challenged with V. anguillarum and WSSV [57]. Nuclear factor NF-kappa-B p110,also known as relish, is an important nuclear transcription factor in the IMD signaling pathway which functions parallel to the Toll pathway [53, 58]. M. rosenbergii relish was reported to be involved in bacterial infection and overexpression of relish induced the expression of various AMPs [59]. In this study, we found that the toll and NF-kappa-B p110 expression were up-regulated after the infection of MrNV, whereas the expression of spätzle was down-regulated. We also validated the expression of spätzle using qPCR which was 3.26-fold down-regulated compared to 17.88-fold down-regulated in RNAseq (Table 5). These results suggested that these genes are involved in the immune system against MrNV infection. Down-regulation of spätzle may be caused by a negative feedback regulation from the activation of the toll pathway [60, 61].
Antimicrobial peptides (AMPs) are also important components in first line of immune system. AMPs are usually small cationic, amphipathic, germ-line encoded proteins that have rapid and efficient antimicrobial effects against broad spectrum of microorganisms including bacteria, fungi, and viruses. AMPs differ in structural conformation, charge, and amphipathicity [62, 63]. Fundamentally, AMPs disrupt the membrane integrity of the target and then destroy the microbe by membrane destabilization or pore formation [62, 64]. Several studies reported that anti-lipopolysaccharide factors (ALFs) and lysozymes expression were up-regulated after the infection of WSSV in M. rosenbergii, F. chinensis, and L. vannamei [35, 65, 66]. Crustin expression in M. rosenbergii hemocytes was found to be up-regulated after WSSV, infectious hypodermal and hematopoietic necrosis virus (IHHNV), and Aeromonas hydrophila challenges [67]. However, crustin isoform 1 and 2 expression in P. monodon were down-regulated after WSSV infection, whereas crustin isoform 3 was up-regulated [68]. In our report, ALFs, i-type lysozyme-like protein 2 (LYZL2), and crustin members were differentially expressed during MrNV infection. ALFs and LYZL2 were up-regulated, whereas several crustin isoforms were both up- and down-regulated. Additionally, the validation by qPCR showed that ALF1 was up-regulated by 5.86-fold which was comparable to the 2.25-fold up-regulation by RNAseq (Table 5). These suggested that ALFs and LYZL2 are involved in the prawn immune response against MrNV, whereas only certain isoforms of crustin are involved.
Blood clotting is a humoral response which prevents hemolymph loss and microbial spread during injury [69]. In crustacean, blood clotting involves cross-linking aggregation of clotting proteins (CPs) by calcium-dependent transglutaminase (TGase) produced by the hemocyte [70]. Previous studies showed that lysozyme and crustin expression in M. japonicus were down-regulated in TGase depleted prawn suggesting that there is a link between the blood clotting system and AMPs [71]. We found that transglutaminase and hemicentin-1-like isoform X2 (HMCN1) expression were up-regulated. In addition, we validated the expression of HMCN1 which was up-regulated by 5.31-fold in the RNAseq and 16.07-fold by qPCR (Table 5). These results suggested that these two blood coagulation components are involved as a response to the infection of MrNV. Moreover, up-regulation of two blood coagulation components, lysozyme, and certain isoforms of crustin may indicate a link between the blood clotting system and AMPs in M. rosenbergii.
Apoptosis or programmed cell death plays an important role in cellular immune response by limiting viral replication and eliminating virus-infected cells in multicellular organisms [16, 72]. Apoptosis requires the activation of caspases, highly conserved cysteine proteases that are involved in the execution of cell death [73]. Previous studies characterized M. rosenbergii caspase (MrCasp) and demonstrated that the capsid protein of MrNV could inhibit apoptotic responses of MrCasp in Sf9 cells [74]. M. rosenbergii caspase 3c expression in hemocyte was found to be up-regulated after IHHNV challenge [75]. M. rosenbergii inhibitor of apoptosis protein (IAP) has been characterized and was found to be up-regulated in hepatopancreas after IHHNV infection [76]. IAP is one of the apoptosis regulators that binds and inhibits the activity of caspase [77]. Additionally, L. vannamei IAPs-silenced by RNAi demonstrated higher expression of WSSV genes and significant down-regulation of AMP genes [35]. In this study, we reported up-regulation of caspase, caspase4, and IAP in response to MrNV challenge. We also validated the expression of caspase which was 2.64-fold up-regulation in RNAseq and 12.05-fold by qPCR (Table 5). Up-regulation of these genes suggests that these gene are involved in MrNV infection.
Phagocytosis plays a role in innate immune system by ingesting microparticles including microbial pathogens and cellular debris from apoptosis and necrosis [78]. Recent studies reported that the small GTPases play a role in antiviral immunity by controlling cellular trafficking and regulating phagocytosis [79,80,81]. ADP ribosylation factors (Arfs), which are small ubiquitously expressed GTPases have been characterized in M. rosenbergii and M. japonicus. Arfs expression was up-regulated in both species after WSSV challenge [82, 83] [84]. The expression of Rab GTPases, Ras-like GTPase superfamily members, are up-regulated in response to WSSV [85] and MrNV infection. We validated, ADP ribosylation factors (Arfs), using qPCR and found 5.45-fold up-regulation compared to 2.19-fold by RNAseq (Table 5). These indicates involvement of these small GTPases in antiviral immunity against MrNV.
Antioxidant enzymes are responsible for removing harmful reactive oxygen species (ROS) during the prawn immune response [86]. Several antioxidant enzymes have been identified in M. rosenbergii such as selenium dependent glutathione peroxidase [36], glutathione S-transferase (GST) [87], copper/zinc superoxide dismutase (CuZnSOD) [88], and thiol-dependent peroxiredoxin (Prdx) [89]. These genes have been reported to be differentially expressed according to different types of pathogen [36, 87,88,89]. We identified five antioxidant enzyme genes that were differentially expressed during MrNV infection. We also validated the expression of one of these genes, CuZnSOD3, using qPCR which was 5.72-fold down-regulated compared to 13.45-fold down-regulated in RNAseq (Table 5).
In prawns, RNA interference (RNAi) plays a crucial role in antiviral immunity. Several key components of RNAi has been characterized in prawns including isoforms of dicer and argonaute in M. rosenbergii [90], L. vannamei [91,92,93], and P. monodon [94, 95]. Dicer, RNase III-like enzyme, is responsible for the cleavage of long dsRNA into 21–30 bp siRNA which initiates RNAi mechanism [96, 97]. Genome derived silencing RNAs (siRNAs) are unwound and incorporated with argonaute protein which is the main component of the RNA-induced silencing complex (RISC) [98]. RISC then recognizes and degrades target mRNA which represses the expression of the target genes [99]. Previous studies reported that administration of synthetic dsRNA/ siRNA specific to genes of yellow head virus (YHV) [100, 101], or WSSV [102] resulted in specific inhibition of viral replication. Moreover, dsRNA can be formed during the replication of both RNA (taura syndrome virus (TSV) and YHV) and DNA viruses (WSSV) which triggers antiviral responses via RNAi [103]. Recent studies revealed that 31 miRNAs were differentially expressed during WSSV infection suggesting that these miRNA may be involved in innate immunity [104]. Additionally, a total of 24 miRNAs have been identified in M. japonicus and reported that these miRNAs may be involved in innate immunity including regulating processes such as phagocytosis, proPO system, and apoptosis [105]. Taken together, both siRNAs and miRNAs have participated in shrimp antiviral immunity. In this study, dicer-2 and argonaute-3 were significantly up-regulated in response to the MrNV infection. Importantly, the qPCR validation showed 6.81-fold up-regulation of dicer-2 compared to 3.66-fold up-regulation from RNAseq results (Table 5) indicating that the production of RNAi may play an antiviral role against MrNV infection in M. rosenbergii.
Conclusion
In conclusion, this study reported a highly complete transcriptome from the post-larvae stage of giant river prawn, M. rosenbergii. This transcriptome expands the transcriptomic resources for further gene functional analysis and transcriptome profiling in response to certain conditions. Transcripts abundance analysis between control and MrNV-infected group revealed significant differences in the transcript abundance of various immune responses e.g. immune signaling pathway, prophenol oxidase system, antimicrobial peptides, blood clotting system, phagocytosis and apoptosis, and RNA interference. To our knowledge, this study is the first report of the transcriptomic profile of M. rosenbergii post-larvae during the infection of MrNV. This study also provides preliminary insight on molecular responses of the prawn to MrNV infection that improve our understanding of M. rosenbergii antiviral responses and may provide molecular targets to help contain the disease outbreak.
Materials and methods
Preparation of MrNV infected M. rosenbergii post-larvae
M. rosenbergii post-larvae (PL 25–30) were purchased from local farm at Suphan Buri provinces, Thailand. The healthy PL were kept in a glass tank with dechlorinated freshwater and continuous aeration at room temperature (25–27 °C). After 1 day of acclimation, the PL were divided into two groups including control and MrNV group. In the MrNV group, the PL were challenged with 10% w/v MrNV-infected PL homogenated in TN buffer (20 mM Tris-HCl and 0.4 M NaCl, pH 7.4) using the immersion method [106]. The control group were challenged with TN buffer instead of MrNV. After 4 days of immersion, the MrNV-infected PL and the PL from control group were divided into 10 subgroups (10 PL per each subgroup). Six subgroups were used in Illumina sequencing, whereas four subgroups were used in quantitative RT-PCR experiment. The PL samples were immersed in DNA/RNA Shield (Zymo Research, USA) and stored at -80 °C until RNA extraction.
The PL from both groups were subjected to MrNV detection using RT-PCR. Viral nucleic acid was extracted from the PL using High Pure Viral Nucleic Acid Kit (Roche). The MrNV-specific primers designed by Senapin and others were used in this study [107]. The RT-PCR was carried out using SuperScript® III One-Step RT-PCR System (Invitrogen) and the extracted viral nucleic acid as a template. The RT-PCR condition was 50 °C for 30 min, followed by 94 °C for 5 min, 35 cycles of 94 °C for 1 min, 55 °C for 45 s, and 72 °C for 1 min, and final extension at 72 °C for 10 min.
RNA extraction and Illumina sequencing
Total RNA was extracted from each subgroup using Quick-RNA™ MiniPrep (Zymo Research, USA) according to manufacturer’s protocol. The extracted RNA was quantified using DropSense 16 Micro-volume spectrophotometer (Unchained Labs, USA) and stored at -80 °C until library preparation.
The cDNA library was prepared using SENSE mRNA-Seq Library Prep Kit V2 (Lexogen, USA) and the purification module with magnetic beads (Lexogen, USA). Each library was indexed by 6-nucleotide-long i7 indices during library amplification step according to manufacturer’s protocol. The prepared cDNA library was subjected to quality assessment using Qubit 4 Fluorometer (Invitrogen, USA) and LabChip GX Touch 24 microfluidic nucleic acid analyzer (PerkinElmer, USA). Before denaturation, equal amount of each cDNA library were pooled together and re-purified using the magnetic beads. Total of 2 nM pooled library was subjected to denaturation steps according to NextSeq System Denature and Dilute Libraries Guide (Illumina, USA). Cluster generation and paired-end sequencing with 75 bp were performed on a NextSeq 550 sequencer (Illumina, USA) using NextSeq 500/550 High Output Reagent Cartridge v2 150 cycles (Illumina, USA).
Data analysis pipeline
The automated data analysis pipeline was written in python using the Snakemake tool [108]. The data analysis pipeline contained three major sections including raw data pre-processing, transcriptome assembly, and post-processing of the transcriptome (Fig. 8). Quality assessment of the sequence data was performed using FastQC v. 0.11.5 [109] and complied using MultiQC v 1.8 [110]. Raw read data were subjected to quality trimming including trim low quality bases, removing N nucleotides, and discarding reads below 36 bases long using Trimmomatic v 0.36 [111]. The trimmed reads were subjected to quality assessment using FastQC and MultiQC and then merged for transcriptome assembly using merge command.
The overviews of analysis pipeline. The pipeline is divided into three major parts including raw data pre-processing, transcriptome assembly, and post-precessing of the transcriptome which indicated by area of different colors. Boxes represent datasets. Rounded boxes represent analyses. The software for each analysis is indicated at the top of the box whereas the database for homology search are listed under the box
The de novo transcriptome assembly was performed using Trinity software v 2.8.0 using default parameters [112]. The in-silico normalization was performed within Trinity prior to de novo assembly. This process resulted in transcripts with relatively high redundancy. To remove the redundancy, transcripts that have more than 95% of identity were clustered together using CD-HIT software [113]. The final transcriptome was subjected to quality assessment including calculate the fragment mapping rates using Bowtie 2 v 2.3.0 [114], examine orthologs completeness using BUSCO v 3 [115] against arthropoda_odb9 database, and generate Nx and ExN50 statistics using ‘contig_ExN50_statistic.pl’ script within Trinity assembler.
The final transcriptome was aligned against UniProt and NCBI’s non-redundant arthropod database using Blastx. The results from Blastx against UNIPROT database were then used to obtain functional annotation from EggNOG, KEGG, and GO database using Trinotate v 3.0.2 [40].
To identify differentially abundant transcripts, the raw read counts were calculated by RSEM software [116] and then used to generate abundance matrix. Differential abundance analysis was performed using EdgeR [117]. EdgeR uses the trimmed mean of M-values normalization method (TMM) to calculate the transcript abundance levels [118] with the Benjamini-Hochberg method for multiple p-value correction [119]. The transcripts that had at least two-fold change with a false discovery rate (FDR or adjusted p-value) less than 0.05 was considered as differentially abundant transcripts.
Quantitative RT-PCR analysis
Nine differentially abundant unigenes were selected for quantitaltive RT-PCR (qRT-PCR) including anti-lipopolysaccharide factor 1 (ALF1), Spatzle (Spz), copper/zinc superoxide dismutase 3 (CuZnSOD3), caspase (CASP), antiviral protein (Anv), dicer (DICER), hemicentin-1-like (HMCN1), ADP ribosylation factors (ARF), and prophenoloxidase (ProPO). In this study, elongation factor1-alpha (EF1-alpha) were used as an internal reference gene [120]. Primers used in the qRT-PCR experiments were listed in Table 6. Primers were subjected to primer efficiency testing using two-fold dilution of cDNA ranging from 200 ng to 12.5 ng. The efficiency of primers was calculated using the following formula “Efficiency (100%) = (10^(-1/ Slope)-1)*100”. Primers with efficiency score between 90 and 110% were used in the qRT-PCR validation.
Total RNA was extracted from separate biological samples using Quick-RNA™ MiniPrep (Zymo Research, USA). The first-strand cDNA was synthesized using SensiFAST™ cDNA Synthesis Kit (Bioline, UK) according to manufacturer’s protocol. The qPCR was carried out using SensiFAST™ SYBR® Lo-ROX Kit (Bioline, UK). The qPCR condition was 95 °C for 2 min, 40 cycles of 95 °C for 5 s, 60 °C for 10 s, and 72 °C for 15 s, followed by melting curve analysis. The delta-delta Ct method was used to calculate relative fold-change of gene expression between control and infected samples [121]. Statistical differences between two groups were conducted using a simple student t-test. The results from qPCR were then compared with the results from RNAseq pipeline using heatmap and coefficient of determination (R2).
Availability of data and materials
Raw data has been uploaded to the National Centre for Biotechnology Information Sequence Read Archive (SRA) under the accession BioProject number: PRJNA550272. The data analysis pipeline is available on GitHub at https://github.com/prawnseq/Mrosenbergii_MrNV_RNAseq. Annotation results and full-list of differentially expressed transcripts are available in Additional file 1: Tables S1 and S2 and Additional file 2: Table S3.
Abbreviations
- AMPs:
-
Antimicrobial peptides
- cDNA:
-
Complementary deoxyribonucleic acid
- CPM:
-
Count per millions
- EggNOG:
-
Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups
- FDR:
-
False discovery rate
- GO:
-
Gene Ontology
- IHHNV:
-
Infectious hypodermal and hematopoietic necrosis virus
- KEGG:
-
Kyoto Encyclopedia of Genes and Genome pathway
- LogFC:
-
Log fold change
- miRNA:
-
Micro ribonucleic acid
- mRNA:
-
Messenger ribonucleic acid
- MrNV:
-
Macrobrachium rosenbergii nodavirus
- NCBI:
-
National Center for Biotechnology Information
- NGS:
-
Next generation sequencing
- PAMPs:
-
Pathogen-associated molecular patterns
- PC1:
-
Principle component 1
- PCA:
-
Principle component analysis
- PL:
-
Post larvae
- ProPO:
-
Prophenol oxidase
- PRRs:
-
Pattern recognition receptors
- qRT-PCR:
-
Quantitative reverse transcription polymerase chain reaction
- RdRp:
-
RNA-dependent RNA polymerase
- RNAi:
-
RNA interference
- RNAseq:
-
Ribonucleic acid sequencing
- siRNA:
-
Small interfering ribonucleic acid
- SRA:
-
Sequence Read Archive
- TMM:
-
Trimmed mean of M-values normalization method
- TPM:
-
Transcript per millions
- WSSV:
-
White spot syndrome virus
- WTD:
-
White tail disease
References
FAO. FAO yearbook. Fishery and aquaculture statistics. 2016. Rome: FAO; 2018.
Lightner DV, Redman RM. Shrimp diseases and current diagnostic methods. Aquaculture. 1998;164(1–4):201–20.
Arcier JM, Herman F, Lightner DV, Redman RM, Mari J, Bonami JR. A viral disease associated with mortalities in hatchery-reared postlarvae of the giant freshwater prawn Macrobrachium rosenbergii. Dis Aquat Organ. 1999;38:177–81.
Tung CW, Wang CS, Chen SN. Histological and electron microscopy study on macrobrachium muscle virus (MMV) infection in the giant freshwater prawn, Macrobrachium rosenbergii (de man), cultured in Taiwan. J Fish Dis. 1999;22:71–5.
Qian D, Shi Z, Zhang S, Cao Z, Liu W, Li L, et al. Extra small virus-like particle (XSV) and nodavirus associated with whitish muscle disease in the giant freshwater prawn, macrobrachium rosenbergii. J Fish Dis. 2003;26:521–7.
Sahul Hameed AS, Yoganandhan K, Sri Widada J, Bonami JR. Experimental transmission and tissue tropism of Macrobrachium rosenbergii nodavirus (MrNV) and its associated extra small virus (XSV). Dis Aquat Organ. 2004;62(3):191–6.
Yoganandhan K, Sri Widada J, Bonami JR, Sahul Hameed AS. Simultaneous detection of Macrobrachium rosenbergii nodavirus and extra small virus by a single tube, one-step multiplex RT-PCR assay. J Fish Dis. 2005;28(2):65–9.
Owens L, La Fauce K, Juntunen K, Hayakijkosol O, Zeng C. Macrobrachium rosenbergii nodavirus disease (white tail disease) in Australia. Dis Aquat Organ. 2009;85(3):175–80.
Widada JS, Bonami JR. Characteristics of the monocistronic genome of extra small virus, a virus-like particle associated with Macrobrachium rosenbergii nodavirus: possible candidate for a new species of satellite virus. J Gen Virol. 2004;85(Pt 3):643–6.
Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296(5571):1319–21.
Bonami JR, Shi Z, Qian D, Sri WJ. White tail disease of the giant freshwater prawn, Macrobrachium rosenbergii: separation of the associated virions and characterization of MrNV as a new type of nodavirus. J Fish Dis. 2005;28(1):23–31.
Romestand B, Bonami JR. A sandwich enzyme linked immunosorbent assay (S-ELISA) for detection of MrNV in the giant freshwater prawn, Macrobrachium rosenbergii (de man). J Fish Dis. 2003;26(2):71–5.
Sri Widada J, Durand S, Cambournac I, Qian D, Shi Z, Dejonghe E, et al. Genome-based detection methods of Macrobrachium rosenbergii nodavirus, a pathogen of the giant freshwater prawn, Macrobrachium rosenbergii dot-blot, in situ hybridization and RT-PCR. J Fish Dis. 2003;26(10):583–90.
Soderhall K, Cerenius L. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr Opin Immunol. 1998;10(1):23–8.
Jiravanichpaisal P, Lee BL, Soderhall K. Cell-mediated immunity in arthropods: hematopoiesis, coagulation, melanization and opsonization. Immunobiology. 2006;211(4):213–36.
Koyama AH, Fukumori T, Fujita M, Irie H, Adachi A. Physiological significance of apoptosis in animal virus infection. Microbes Infect. 2000;2(9):1111–7.
Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92(5):255–64.
Greenwood MP, Mecawi AS, Hoe SZ, Mustafa MR, Johnson KR, Al-Mahmoud GA, et al. A comparison of physiological and transcriptome responses to water deprivation and salt loading in the rat supraoptic nucleus. Am J Physiol Regul Integr Comp Physiol. 2015;308(7):R559–68.
Nguyen C, Nguyen TG, Nguyen LV, Pham HQ, Nguyen TH, Pham HT, et al. De novo assembly and transcriptome characterization of major growth-related genes in various tissues of Penaeus monodon. Aquaculture. 2016;464:545–53.
Huerlimann R, Wade NM, Gordon L, Montenegro JD, Goodall J, McWilliam S, et al. De novo assembly, characterization, functional annotation and expression patterns of the black tiger shrimp (Penaeus monodon) transcriptome. Sci Rep. 2018;8(1):13553.
Soonthornchai W, Chaiyapechara S, Klinbunga S, Thongda W, Tangphatsornruang S, Yoocha T, et al. Differentially expressed transcripts in stomach of Penaeus monodon in response to AHPND infection. Dev Comp Immunol. 2016;65:53–63.
Yu Y, Wei J, Zhang X, Liu J, Liu C, Li F, et al. SNP discovery in the transcriptome of white Pacific shrimp Litopenaeus vannamei by next generation sequencing. PLoS One. 2014;9(1):e87218.
Chen K, Li E, Li T, Xu C, Wang X, Lin H, et al. Transcriptome and molecular pathway analysis of the Hepatopancreas in the Pacific white shrimp Litopenaeus vannamei under chronic low-salinity stress. PLoS One. 2015;10(7):e0131503.
Li C, Weng S, Chen Y, Yu X, Lu L, Zhang H, et al. Analysis of Litopenaeus vannamei transcriptome using the next-generation DNA sequencing technique. PLoS One. 2012;7(10):1–12.
Li S, Zhang X, Sun Z, Li F, Xiang J. Transcriptome analysis on Chinese shrimp Fenneropenaeus chinensis during WSSV acute infection. PLoS One. 2013;8(3):e58627.
Shi X, Meng X, Kong J, Luan S, Luo K, Cao B, et al. Transcriptome analysis of 'Huanghai no. 2′ Fenneropenaeus chinensis response to WSSV using RNA-seq. Fish Shellfish Immunol. 2018;75:132–8.
Powell D, Knibb W, Remilton C, Elizur A. De-novo transcriptome analysis of the banana shrimp (Fenneropenaeus merguiensis) and identification of genes associated with reproduction and development. Mar Genomics. 2015;22:71–8.
Wang W, Yang S, Wang C, Shi L, Guo H, Chan S. Gill transcriptomes reveal involvement of cytoskeleton remodeling and immune defense in ammonia stress response in the banana shrimp Fenneropenaeus merguiensis. Fish Shellfish Immunol. 2017;71:319–28.
Sellars MJ, Trewin C, McWilliam SM, Glaves RS, Hertzler PL. Transcriptome profiles of Penaeus (Marsupenaeus) japonicus animal and vegetal half-embryos: identification of sex determination, germ line, mesoderm, and other developmental genes. Mar Biotechnol (NY). 2015;17(3):252–65.
Rao R, Bing Zhu Y, Alinejad T, Tiruvayipati S, Lin Thong K, Wang J, et al. RNA-seq analysis of Macrobrachium rosenbergii hepatopancreas in response to Vibrio parahaemolyticus infection. Gut Pathog. 2015;7:6.
Rao R, Bhassu S, Bing RZ, Alinejad T, Hassan SS, Wang J. A transcriptome study on Macrobrachium rosenbergii hepatopancreas experimentally challenged with white spot syndrome virus (WSSV). J Invertebr Pathol. 2016;136:10–22.
Cao J, Wu L, Jin M, Li T, Hui K, Ren Q. Transcriptome profiling of the Macrobrachium rosenbergii lymphoid organ under the white spot syndrome virus challenge. Fish Shellfish Immunol. 2017;67:27–39.
Ding Z, Jin M, Ren Q. Transcriptome analysis of Macrobrachium rosenbergii intestines under the white spot syndrome virus and poly (I:C) challenges. PLoS One. 2018;13(9):e0204626.
Jariyapong P, Pudgerd A, Cheloh N, Hirono I, Kondo H, Vanichviriyakit R, et al. Hematopoietic tissue of Macrobrachium rosenbergii plays dual roles as a source of hemocyte hematopoiesis and as a defensive mechanism against Macrobrachium rosenbergii nodavirus infection. Fish Shellfish Immunol. 2019;86:756–63.
Wang PH, Wan DH, Gu ZH, Qiu W, Chen YG, Weng SP, et al. Analysis of expression, cellular localization, and function of three inhibitors of apoptosis (IAPs) from Litopenaeus vannamei during WSSV infection and in regulation of antimicrobial peptide genes (AMPs). PLoS One. 2013;8(8):e72592.
Yeh SP, Liu KF, Chiu ST, Jian SJ, Cheng W, Liu CH. Identification and cloning of a selenium dependent glutathione peroxidase from giant freshwater prawn, Macrobrachium rosenbergii. Fish Shellfish Immunol. 2009;27(2):181–91.
Ghaffari N, Sanchez-Flores A, Doan R, Garcia-Orozco KD, Chen PL, Ochoa-Leyva A, et al. Novel transcriptome assembly and improved annotation of the whiteleg shrimp (Litopenaeus vannamei), a dominant crustacean in global seafood mariculture. Sci Rep. 2014;4:7081.
UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D15.
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512.
Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, et al. A tissue-mapped axolotl De novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017;18(3):762–76.
Griffith M, Griffith OL, Mwenifumbo J, Goya R, Morrissy AS, Morin RD, et al. Alternative expression analysis by RNA sequencing. Nat Methods. 2010;7(10):843–7.
Asmann YW, Klee EW, Thompson EA, Perez EA, Middha S, Oberg AL, et al. 3′ tag digital gene expression profiling of human brain and universal reference RNA using Illumina genome analyzer. BMC Genomics. 2009;10:531.
Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11(1):41–6.
Medzhitov R, Janeway C Jr. Innate immunity. N Engl J Med. 2000;343(5):338–44.
Wang XW, Wang JX. Pattern recognition receptors acting in innate immune system of shrimp against pathogen infections. Fish Shellfish Immunol. 2013;34(4):981–9.
Huang X, Huang Y, Shi YR, Ren Q, Wang W. Function of a novel C-type lectin with two CRD domains from Macrobrachium rosenbergii in innate immunity. Dev Comp Immunol. 2015;49(1):121–6.
Zhang XW, Wang XW, Huang Y, Hui KM, Shi YR, Wang W, et al. Cloning and characterization of two different ficolins from the giant freshwater prawn Macrobrachium rosenbergii. Dev Comp Immunol. 2014;44(2):359–69.
Arockiaraj J, Chaurasia MK, Kumaresan V, Palanisamy R, Harikrishnan R, Pasupuleti M, et al. Macrobrachium rosenbergii mannose binding lectin: synthesis of MrMBL-N20 and MrMBL-C16 peptides and their antimicrobial characterization, bioinformatics and relative gene expression analysis. Fish Shellfish Immunol. 2015;43(2):364–74.
Soderhall K, Cerenius L, Johansson MW. The prophenoloxidase activating system and its role in invertebrate defence. Ann N Y Acad Sci. 1994;712:155–61.
Cerenius L, Soderhall K. The prophenoloxidase-activating system in invertebrates. Immunol Rev. 2004;198:116–26.
De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The toll and Imd pathways are the major regulators of the immune response in drosophila. EMBO J. 2002;21(11):2568–79.
Wang PH, Liang JP, Gu ZH, Wan DH, Weng SP, Yu XQ, et al. Molecular cloning, characterization and expression analysis of two novel tolls (LvToll2 and LvToll3) and three putative Spatzle-like toll ligands (LvSpz1-3) from Litopenaeus vannamei. Dev Comp Immunol. 2012;36(2):359–71.
Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743.
Srisuk C, Longyant S, Senapin S, Sithigorngul P, Chaivisuthangkura P. Molecular cloning and characterization of a toll receptor gene from Macrobrachium rosenbergii. Fish Shellfish Immunol. 2014;36(2):552–62.
Feng J, Zhao L, Jin M, Li T, Wu L, Chen Y, et al. Toll receptor response to white spot syndrome virus challenge in giant freshwater prawns (Macrobrachium rosenbergii). Fish Shellfish Immunol. 2016;57:148–59.
Vaniksampanna A, Longyant S, Charoensapsri W, Sithigorngul P, Chaivisuthangkura P. Molecular isolation and characterization of a spatzle gene from Macrobrachium rosenbergii. Fish Shellfish Immunol. 2019;84:441–50.
Shi XZ, Zhang RR, Jia YP, Zhao XF, Yu XQ, Wang JX. Identification and molecular characterization of a Spatzle-like protein from Chinese shrimp (Fenneropenaeus chinensis). Fish Shellfish Immunol. 2009;27(5):610–7.
Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, et al. Relish, a central factor in the control of humoral but not cellular immunity in drosophila. Mol Cell. 1999;4(5):827–37.
Shi YR, Jin M, Ma FT, Huang Y, Huang X, Feng JL, et al. Involvement of relish gene from Macrobrachium rosenbergii in the expression of anti-microbial peptides. Dev Comp Immunol. 2015;52(2):236–44.
Misra S, Hecht P, Maeda R, Anderson KV. Positive and negative regulation of Easter, a member of the serine protease family that controls dorsal-ventral patterning in the drosophila embryo. Development. 1998;125(7):1261–7.
Towb P, Bergmann A, Wasserman SA. The protein kinase Pelle mediates feedback regulation in the drosophila toll signaling pathway. Development. 2001;128(23):4729–36.
Yount NY, Bayer AS, Xiong YQ, Yeaman MR. Advances in antimicrobial peptide immunobiology. Biopolymers. 2006;84(5):435–58.
Bulet P, Cociancich S, Dimarcq JL, Lambert J, Reichhart JM, Hoffmann D, et al. Insect immunity. Isolation from a coleopteran insect of a novel inducible antibacterial peptide and of new members of the insect defensin family. J Biol Chem. 1991;266(36):24520–5.
Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–50.
Ren Q, Zhang Z, Li XC, Jie D, Hui KM, Zhang CY, et al. Three different anti-lipopolysaccharide factors identified from giant freshwater prawn, Macrobrachium rosenbergii. Fish Shellfish Immunol. 2012;33(4):766–74.
Wang B, Li F, Dong B, Zhang X, Zhang C, Xiang J. Discovery of the genes in response to white spot syndrome virus (WSSV) infection in Fenneropenaeus chinensis through cDNA microarray. Mar Biotechnol (NY). 2006;8(5):491–500.
Arockiaraj J, Gnanam AJ, Muthukrishnan D, Gudimella R, Milton J, Singh A, et al. Crustin, a WAP domain containing antimicrobial peptide from freshwater prawn Macrobrachium rosenbergii: immune characterization. Fish Shellfish Immunol. 2013;34(1):109–18.
Swapna PA, Rosamma P, Valsamma J, BrightSingh IS. Anti-lipopolysaccharide factor and crustin-III, the anti-white spot virus peptides in Penaeus monodon: control of viral infection by up-regulation. Aquaculture. 2011;319(1–2):11–7.
Maningas MB, Kondo H, Hirono I. Molecular mechanisms of the shrimp clotting system. Fish Shellfish Immunol. 2013;34(4):968–72.
Hall M, Wang R, van Antwerpen R, Sottrup-Jensen L, Soderhall K. The crayfish plasma clotting protein: a vitellogenin-related protein responsible for clot formation in crustacean blood. Proc Natl Acad Sci U S A. 1999;96(5):1965–70.
Fagutao FF, Maningas MB, Kondo H, Aoki T, Hirono I. Transglutaminase regulates immune-related genes in shrimp. Fish Shellfish Immunol. 2012;32(5):711–5.
Everett H, McFadden G. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 1999;7(4):160–5.
Menze MA, Fortner G, Nag S, Hand SC. Mechanisms of apoptosis in Crustacea: what conditions induce versus suppress cell death? Apoptosis. 2010;15(3):293–312.
Youngcharoen S, Senapin S, Lertwimol T, Longyant S, Sithigorngul P, Flegel TW, et al. Interaction study of a novel Macrobrachium rosenbergii effector caspase with B2 and capsid proteins of M. rosenbergii nodavirus reveals their roles in apoptosis. Fish Shellfish Immunol. 2015;45(2):534–42.
Arockiaraj J, Easwvaran S, Vanaraja P, Singh A, Othman RY, Bhassu S. Effect of infectious hypodermal and haematopoietic necrosis virus (IHHNV) infection on caspase 3c expression and activity in freshwater prawn Macrobrachium rosenbergii. Fish Shellfish Immunol. 2012;32(1):161–9.
Arockiaraj J, Vanaraja P, Easwvaran S, Singh A, Othman RY, Bhassu S. Bioinformatic characterization and gene expression pattern of apoptosis inhibitor from Macrobrachium rosenbergii challenged with infectious hypodermal and hematopoietic necrosis virus. Fish Shellfish Immunol. 2011;31(6):1259–67.
Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 1997;16(23):6914–25.
Stuart LM, Ezekowitz RA. Phagocytosis and comparative innate immunity: learning on the fly. Nat Rev Immunol. 2008;8(2):131–41.
Liu W, Han F, Zhang X. Ran GTPase regulates hemocytic phagocytosis of shrimp by interaction with myosin. J Proteome Res. 2009;8(3):1198–206.
Myers KR, Casanova JE. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 2008;18(4):184–92.
Wu W, Zong R, Xu J, Zhang X. Antiviral phagocytosis is regulated by a novel Rab-dependent complex in shrimp penaeus japonicus. J Proteome Res. 2008;7(1):424–31.
Ding ZF, Ren J, Tan JM, Wang Z, Yin SW, Huang Y, et al. Characterization of two novel ADP ribosylation factors from giant freshwater prawn Macrobrachium rosenbergii and their responses to WSSV challenge. Dev Comp Immunol. 2015;48(1):204–9.
Zhang M, Ma J, Lei K, Xu X. Molecular cloning and characterization of a class II ADP ribosylation factor from the shrimp Marsupenaeus japonicus. Fish Shellfish Immunol. 2010;28(1):128–33.
Ma J, Zhang M, Ruan L, Shi H, Xu X. Characterization of two novel ADP ribosylation factors from the shrimp Marsupenaeus japonicus. Fish Shellfish Immunol. 2010;29(6):956–62.
Huang Y, Ren Q. Identification and function of 11 Rab GTPases in giant freshwater prawn Macrobrachium rosenbergii. Fish Shellfish Immunol. 2015;43(1):120–30.
De la Fuente M, Victor VM. Anti-oxidants as modulators of immune function. Immunol Cell Biol. 2000;78(1):49–54.
Arockiaraj J, Gnanam AJ, Palanisamy R, Bhatt P, Kumaresan V, Chaurasia MK, et al. A cytosolic glutathione s-transferase, GST-theta from freshwater prawn Macrobrachium rosenbergii: molecular and biochemical properties. Gene. 2014;546(2):437–42.
Cheng W, Tung YH, Liu CH, Chen JC. Molecular cloning and characterisation of copper/zinc superoxide dismutase (cu,Zn-SOD) from the giant freshwater prawn Macrobrachium rosenbergii. Fish Shellfish Immunol. 2006;21(1):102–12.
Arockiaraj J, Easwvaran S, Vanaraja P, Singh A, Othman RY, Bhassu S. Immunological role of thiol-dependent peroxiredoxin gene in Macrobrachium rosenbergii. Fish Shellfish Immunol. 2012;33(1):121–9.
Shpak N, Manor R, Abilevich LK, Mantal O, Shavit K, Aflalo ED, et al. Short versus long double-stranded RNA activation of a post-transcriptional gene knockdown pathway. RNA Biol. 2017;14(12):1766–75.
Yao X, Wang L, Song L, Zhang H, Dong C, Zhang Y, et al. A Dicer-1 gene from white shrimp Litopenaeus vannamei: expression pattern in the processes of immune response and larval development. Fish Shellfish Immunol. 2010;29(4):565–70.
Chen YH, Jia XT, Zhao L, Li CZ, Zhang S, Chen YG, et al. Identification and functional characterization of Dicer2 and five single VWC domain proteins of Litopenaeus vannamei. Dev Comp Immunol. 2011;35(6):661–71.
Labreuche Y, Veloso A, de la Vega E, Gross PS, Chapman RW, Browdy CL, et al. Non-specific activation of antiviral immunity and induction of RNA interference may engage the same pathway in the Pacific white leg shrimp Litopenaeus vannamei. Dev Comp Immunol. 2010;34(11):1209–18.
Su J, Oanh DT, Lyons RE, Leeton L, van Hulten MC, Tan SH, et al. A key gene of the RNA interference pathway in the black tiger shrimp, Penaeus monodon: identification and functional characterisation of Dicer-1. Fish Shellfish Immunol. 2008;24(2):223–33.
Unajak S, Boonsaeng V, Jitrapakdee S. Isolation and characterization of cDNA encoding Argonaute, a component of RNA silencing in shrimp (Penaeus monodon). Comp Biochem Physiol B Biochem Mol Biol. 2006;145(2):179–87.
Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15(2):188–200.
Hannon GJ. RNA interference. Nature. 2002;418(6894):244–51.
Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4(6):457–67.
Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–6.
Yodmuang S, Tirasophon W, Roshorm Y, Chinnirunvong W, Panyim S. YHV-protease dsRNA inhibits YHV replication in Penaeus monodon and prevents mortality. Biochem Biophys Res Commun. 2006;341(2):351–6.
Tirasophon W, Roshorm Y, Panyim S. Silencing of yellow head virus replication in penaeid shrimp cells by dsRNA. Biochem Biophys Res Commun. 2005;334(1):102–7.
Xu J, Han F, Zhang X. Silencing shrimp white spot syndrome virus (WSSV) genes by siRNA. Antiviral Res. 2007;73(2):126–31.
Huang T, Zhang X. Host defense against DNA virus infection in shrimp is mediated by the siRNA pathway. Eur J Immunol. 2013;43(1):137–46.
Huang T, Zhang X. Functional analysis of a crustacean microRNA in host-virus interactions. J Virol. 2012;86(23):12997–3004.
Yang G, Yang L, Zhao Z, Wang J, Zhang X. Signature miRNAs involved in the innate immunity of invertebrates. PLoS One. 2012;7(6):e39015.
Sahoo PK, Shekhar MS, Das A, Kumar MD, Pillai BR, Sahul HA. Immunomodulatory effect of recombinant RNA-dependent RNA polymerase protein of Macrobrachium rosenbergii nodavirus in giant freshwater prawn M. rosenbergii. Aquacult Res. 2012;43(8):1096–106.
Senapin S, Phiwsaiya K, Gangnonngiw W, Briggs M, Sithigorngul P, Flegel TW. Infections of MrNV (Macrobrachium rosenbergii nodavirus) in cultivated whiteleg shrimp Penaeus vannamei in Asia. Aquaculture. 2012;338–341:41–6.
Koster J, Rahmann S. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics. 2012;28(19):2520–2.
Andrew S. FastQC: a quality control tool for high throughput sequence data 2010 [Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc]. Accessed 18 Apr 2019.
Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–8.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52.
Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–9.
Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–9.
Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–2.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300.
Dhar AK, Bowers RM, Licon KS, Veazey G, Read B. Validation of reference genes for quantitative measurement of immune gene expression in shrimp. Mol Immunol. 2009;46(8–9):1688–95.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–8.
Acknowledgements
Not applicable.
Funding
This work was financially supported by Strategic Wisdom and Research Institute, Srinakharinwirot University Grant 520/2561 to PC, Science Achievement Scholarship of Thailand to PP, and Natural Sciences and Engineering Research Council of Canada Discovery Grant RGPIN36481–08 to WGB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
PC and WGB conceived and designed the study. PP performed the experiments. PP, CH, PS, SL, WGB, and PC analyzed the data. PC, SL, and PS supervised the sample preparations. PC, WGB, and CH supervised the next-generation sequencing experiments. PP drafted the manuscript. PC, WGB, and CH revised the final draft. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Annotation results of assembled transcriptome against UniProt database. Table S2. Annotation results of assembled transcriptome against Non-redundant Arthropod database.
Additional file 2: Table S3.
Full-list of differentially expressed transcripts using EdgeR.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Pasookhush, P., Hindmarch, C., Sithigorngul, P. et al. Transcriptomic analysis of Macrobrachium rosenbergii (giant fresh water prawn) post-larvae in response to M. rosenbergii nodavirus (MrNV) infection: de novo assembly and functional annotation. BMC Genomics 20, 762 (2019). https://doi.org/10.1186/s12864-019-6102-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-6102-6