Abstract
Background
Pseudocohnilembus persalinus, a unicellular ciliated protozoan, is one of commonest facultative pathogens. We sequenced the macronuclear genome of P. persalinus in 2015, which provided new insights into its pathogenicity.
Results
Here, we present the P. persalinus genome database (PPGD) (http://ciliates.ihb.ac.cn/database/home/#pp), the first genome database for the scuticociliatosis pathogens. PPGD integrates P. persalinus macronuclear genomic and transcriptomic data, including genome sequence, transcript, gene expression data, and gene annotation, as well as relevant information on its biology, morphology and taxonomy. The database also provides functions for visualizing, analyzing, and downloading the data.
Conclusion
PPGD is a useful resource for studying scuticociliates or scuticociliatosis. We will continue to update the PPGD by integrating more data and aim to integrate the PPGD with other ciliate databases to build a comprehensive ciliate genome database.
Similar content being viewed by others
Background
The Pseudocohnilembus persalinus scuticociliate is a free-living marine ciliate first reported by Evans and Thompson in 1964 [1]. Since then, its morphological, ecological, and phylogenetic have been well studied [2,3,4]. Since Kim and colleagues (2004) isolated P. persalinus from a diseased olive flounder in Korea, the organism has been recognized as a common facultative pathogen. P. persalinus infection causes scuticociliatosis, one of the most important fish diseases that has led to serious economic losses in marine aquaculture worldwide [5, 6]. P. persalinus is a facultative parasite and can be easily grown in the laboratory by feeding with bacteria. It is therefore a suitable model for studying the life cycle, genetics, and genomics of ciliates. Therefore, P. persalinus is an ideal model for investigating scuticociliatosis and the molecular mechanisms of its pathogenicity.
Like other ciliates, it has two types of functionally diverse nuclei within the same cytoplasm: a micronucleus (MIC) and a macronucleus (MAC). The MIC is transcriptionally silent and undergoes meiosis and transmits the genetic information to the progeny by sexual reproduction. In contrast, the MAC is highly polyploid and transcriptionally active, and controls the non-reproductive features of cell function. In 2015, we reported the P. persalinus MAC genome sequence [7]. This was the first report of its type for a scuticociliate and a marine ciliate. Before that, the genomes of Tetrahymena thermophila, Paramecium tetraurelia and Ichthyophthirius multifiliis have first been sequenced and their comparative genomics analyses have comprehensively provided a better understanding of the unique features between the free-living (T. thermophila and P. tetraurelia) and typical parasitic (I. multifiliis) ciliates. As has been done for these ciliates, we have constructed the P. persalinus genome database (PPGD), which integrates genomic data, transcriptomic data, and gene annotation, thereby providing a useful resource for studying scuticociliates or scuticociliatosis.
Construction
Figure 1 shows a schematic diagram of the structure of the PPGD. Genomic sequence data, transcriptomic sequence (RNA-Seq) data, and genome annotation data are stored in the MySQL database. To use BLAST tool, genomic sequence was also formatted as BLAST database. All data are freely available for downloading. The user-friendly interface and web pages were developed with PHP and HTML. The entire PPGD runs on an Apache HTTP server and uses MySQL as the database management system. In addition, the Generic Genome Browser (GBrowse) [8], a mature, widely used genome browser, was incorporated to manipulate and visualize genome annotation and expression data. Online analysis tools in PPGD were implemented with in-house PHP scripts or by wrapping existing third-party tools (e.g. BLAST server).
Content
PPGD mainly contains three types of datasets containing: (i) genomic sequence; (ii) gene expression data; and (iii) genome annotation. The PPGD also includes a brief summary of the biological, morphological, and taxonomical characteristics of P. persalinus.
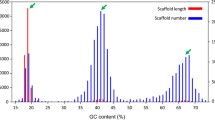
The P. persalinus genome was sequenced with the Illumina platform and then assembled with SOAPdenovo [9]. The 55.5 Mb genome was assembled from 288 scaffolds; the scaffold N50 is approximately 368 kb [7]. A total of 13,186 genes were predicted by an ab initio prediction pipeline. Three types of annotation were included for these genes. The first type was functional prediction based on the BLAST hits when searched against the NCBI non-redundant protein database. The function of 4,265 genes (32%) could be assigned in this way; all other genes were annotated as “hypothetical protein”. The second type was protein domain or structure annotation based on InterProScan prediction [10]. InterProScan integrates the results from multiple protein domain databases (Pfam, PRINTS, PANTHER, Gene3D and InterProScan) and some protein domain structures (coiled-coil, signal peptide, and transmembrane helix). A total of 11,948 genes could be annotated and 5,869 genes could be assigned GO (gene ontology) numbers based on InterProScan predictions. The third type was based on gene information on homologs in other ciliates. Ortholog groups in six ciliates (Ichthyophthirius multifiliis, Oxytricha trifallax, Paramecium tetraurelia, P. persalinus, Stylonychia lemnae, and Tetrahymena thermophila) were obtained using OrthoMCL [11], finally, 5,962 P. persalinus genes could be assigned, and thus provided in PPGD. We also used RNA-Seq data to obtain gene expression data for P. persalinus fed with bacteria to determine changes in gene transcription level under growth condition. The RNA-seq data was mapped to the P. persalinus MAC assembly using TopHat2 [12], then raw read counts were calculated for each gene using Subread [13] program, and the gene expression values were obtained by normalized the raw read counts to RPKM (reads per kilobase per million mapped reads) values. For all the 13,186 predicted genes, 92% (12,145) of genes have more or less reads mapped in the growth condition. Among them, 56% (6765) of genes showed an expression value larger than 10.
Utility and discussion
Search functions
PPGD can be accessed via an easy-to-use web interface. The top row of navigation tabs (including “HOME,” “SEARCH,” “GENOME BROWSER,” “BLAST,” “MORPHOLOGY,” and “DATA DOWNLOAD”) directs the user to retrieve the information. The “HOME” page displays some background information on P. persalinus, introduces the database, and contains news and references related to the PPGD.
A search box (located at the top right of the web page; Fig. 2a) enables the user to search for “Gene ID,” “Keywords,” or “Scaffold ID” in the database (Fig. 2a). We recommend searching the database using a “Gene ID”. The search hits are listed and hyperlinks direct the user to detailed information about the gene. This consists of five main sections (Fig. 3): (i) basic information, including the gene name and location (Fig. 3a); (ii) a GBrowse snapshot showing the gene structure (Fig. 3b); (iii) protein domain, protein structure and GO annotation (Fig. 3c, d); (iv) gene information on homolog (Fig. 3e); and (v) coding sequence (CDS) and protein sequence (Fig. 3f).
The PPGD also offers a search function for homologous sequence in BLAST, which is embedded into the database to provide a graphical interface (Fig. 2b). This allows users to search for gene information on homologs by directly inserting a query sequence into the text box. The CDS, protein sequence, and whole draft genome sequence of P. persalinus are organized as datasets for BLAST searching. Furthermore, “Expect value” and “Number alignments” options are included as advanced settings to filter low-comparability sequence. Finally, users can select an output format for downloading the homologous sequence.
Data visualization
GBrowse has been implemented in PPGD for visualizing gene annotation and transcriptomic data. Typically, the user can use “Gene ID” or a scaffold region to search within GBrowse (Fig. 4a). The GBrowse search will mainly return three tracks: (i) a putative gene model; (ii) an RNA-Seq coverage plot for growth conditions; and (iii) RPKM values (Fig. 4b). GBrowse provides the user with information on the gene structure and expression levels. In the gene model track, a hyperlink forwards the user to a web page with more detailed gene information. In addition, once an interesting gene or genomic region is selected, the user can scroll and zoom using the navigation bar (shown in the upper right of Fig. 4). The sequence can also be retrieved in FASTA format by selecting the “Download Decorated FASTA File” option in the drop-down menu in GBrowse.
Search result page shows detailed genetic information for the example gene, PPERSA_00127420. a Basic description. b GBrowse snapshot. c InterproScan annotations. (d) Protein domain annotations from various databases. e Ortholog information. f Sequence information, including predicted CDS and protein sequence
Other functions
The “MORPHOLOGY” page of PPGD displays detailed morphological information and the taxonomic status of P. persalinus, based on previous studies [5, 14,15,16,17,18]. Users can also download P. persalinus genomic sequence, protein sequence, and CDS in FASTA format and gene annotation information in CSV format via the “DATA DOWNLOAD” page.
Further development
We will continue to update PPGD by integrating more data, such as functional genomics data. As more and more ciliate genomes are sequenced, we will try to integrate these into PPGD to build a comprehensive genome database for ciliates.
Conclusions
PPGD is the first genome database for a scuticociliate. It integrates basic genomic information of P. persalinus and provides a useful resource for studying scuticociliates and scuticociliatosis.
Abbreviations
- BLAST:
-
Basic Local Alignment Search Tool
- CDS:
-
Coding sequence
- GBrowse:
-
Generic Genome Browser
- GO:
-
Gene Ontology
- MAC:
-
Macronucleus
- MIC:
-
Micronucleus
- NCBI:
-
National Center for Biotechnology Information
- PPGD:
-
Pseudocohnilembus persalinus genome database
- RPKM:
-
Reads per kilobase per million mapped reads
References
Evans FR, Thompson JC Jr. Pseudocohnilembidae n. Fam., a hymenostome ciliate family containing one genus, Pseudocohnilembus n. G., with three new species. J Protozool. 1964;11(3):344–52.
Evans FR, Corliss JO. Morphogenesis in the hymenostome ciliate Pseudocohnilembus persalinus and its taxonomic and phylogenetic implications. J Protozool. 1964;11(3):357–70.
Pomp R, Wilbert N. Taxonomic and ecological studies of ciliates from Australian saline soils: colpodids and hymenostomate ciliates. Aust J Mar Freshwat Res. 1988;39(4):479–95.
Song W-B. Morphological and taxonomical studies on some marine scuticociliates from China Sea, with description of two new species, Philasterides armatalis sp. n. And Cyclidium varibonneti sp. n. (Protozoa: Ciliophora: Scuticociliatida). Acta Protozool. 2000;39:295–322.
Kim SM, Cho JB, Lee EH, Kwon SR, Kim SK, Nam YK, et al. Pseudocohnilembus persalinus (Ciliophora : Scuticociitida) is an additional species causing scuticociliatosis in olive flounder Paralichthys olivaceus. Dis Aquat Org. 2004;62(3):239–44.
Cheung PJ, Nigrelli RF, Ruggieri GD. Studies on the morphology of Uronema marinum Dujardin (Ciliatea: Uronematidae) with a description of the histopathology of the infection in marine fishes. J Fish Dis. 1980;3(4):295–303.
Xiong J, Wang G-Y, Cheng J, Tian M, Pan X-M, Warren A, et al. Genome of the facultative scuticociliatosis pathogen Pseudocohnilembus persalinus provides insight into its virulence through horizontal gene transfer. Sci Rep. 2015;5:1–12.
Stein LD, Mungall C, Shu S-Q, Caudy M, Mangone M, Day A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12(10):1599–610.
Li R-Q, Zhu H-M, Ruan J, Qian W-B, Fang X-D, Shi Z-B, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–72.
Jones P, Binns D, Chang HY, Fraser M, Li W-Z, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–40.
Li L, Stoeckert CJ Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36.
Liao Y, Smyth GK, Shi W. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108.
Xu K-D, Song W-B. Study on some pathogenetic ciliates from the mantle cavity of marine molluscs. China J Fish Sci. 1999;6(2):41–5.
Pan X-M, Ma H-G, Shao C, Lin X-F, Hu X-Z. Stomatogenesis and morphological redescription of Pseudocohnilembus persalinus (Ciliophora: Scuticociliatida). Acta Hydrobiol Sin. 2012;36(3):489–95.
Zhan Z-F, Stoeck T, Dunthorn M, Xu K-D. Identification of the pathogenic ciliate Pseudocohnilembus persalinus (Oligohymenophorea: Scuticociliatia) by fluorescence in situ hybridization. European J Protistol. 2014;50(1):16–24.
Gao F, Warren A, Zhang Q-Q, Gong J, Miao M, Sun P, et al. The all-data-based evolutionary hypothesis of ciliated protists with a revised classification of the phylum Ciliophora (eukaryota, Alveolata). Sci Rep. 2016;6:2045–322.
Feng J-M, Jiang C-Q, Warren A, Tian M, Cheng J, Liu G-L, et al. Phylogenomic analyses reveal subclass Scuticociliatia as the sister group of subclass Hymenostomatia within class Oligohymenophorea. Mol Phylogenet Evol. 2015;90:104–11.
Acknowledgements
We thank Ying Zhu (Nextomics Biosciences Co., Ltd.) for his help and suggestion on constructing the database.
Funding
This project was supported by grants from National Natural Science Foundation of China (No. 31525021) to WM, National Natural Science Foundation of China (No. 31672281) to JX, Knowledge Innovation Program of the Chinese Academy of Sciences to JX, Youth Innovation Promotion Association of the Chinese Academy of Sciences to JX.
Availability of data and materials
The database is freely available at http://ciliates.ihb.ac.cn/database/home/#pp.
Author information
Authors and Affiliations
Contributions
JX, WM and WY conceived the work. KC, WW, JX, WY, and WM constructed the database. WW and KC wrote the manuscript. All authors have approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicate.
Consent for publication
Not applicate.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wei, W., Chen, K., Miao, W. et al. Pseudocohnilembus persalinus genome database - the first genome database of facultative scuticociliatosis pathogens. BMC Genomics 19, 676 (2018). https://doi.org/10.1186/s12864-018-5046-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-018-5046-6