Abstract
Background
The maternally inherited endosymbiont Wolbachia is widespread in arthropods and nematodes and can play an important role in the ecology and evolution of its host through reproductive manipulation. Here, we survey Wolbachia in Belonocnema treatae, a widely distributed North American cynipid gall forming wasp that exhibits regional host specialization on three species of oaks and alternation of sexually and asexually reproducing generations. We investigated whether patterns of Wolbachia infection and diversity in B. treatae are associated with the insect’s geographic distribution, host plant association, life cycle, and mitochondrial evolutionary history.
Results
Screening of 463 individuals from 23 populations including sexual and asexual generations from all three host plants across the southern U.S. showed an average infection rate of 56% with three common Wolbachia strains: wTre1–3 and an additional rare variant wTre4. Phylogenetic analysis based on wsp showed that these strains are unrelated and likely independently inherited. We found no difference in Wolbachia infection frequency among host plant associated populations or between the asexual and sexual generations, or between males and females of the sexual generation. Partially incomplete Wolbachia transmission rates might explain the occurrence of uninfected individuals. A parallel analysis of the mitochondrial cytochrome oxidase I gene in B. treatae showed high mtDNA haplotype diversity in both infected and uninfected populations suggesting an ancestral infection by Wolbachia as well as a clear split between eastern and western B. treatae mtDNA clades with a sequence divergence of > 6%. The strain wTre1 was present almost exclusively in the western clade while wTre2 and wTre3 occur almost exclusively in eastern populations. In contrast, the same strains co-occur as double-infections in Georgia and triple-infections in two populations in central Florida.
Conclusions
The diversity of Wolbachia across geographically and genetically distinct populations of B. treatae and the co-occurrence of the same strains within three populations highlights the complex infection dynamics in this system. Moreover, the association of distinct Wolbachia strains with mitochondrial haplotypes of its host in populations infected by different Wolbachia strains suggests a potential role of the endosymbiont in reproductive isolation in B. treatae.
Similar content being viewed by others
Background
Wolbachia are maternally inherited intracellular bacteria reported in a diverse range of arthropod species worldwide (reviewed in [1]). This endosymbiont has wide reaching impacts on the biology of its host [2], including effects on survival through immunity and nutrition [3, 4], and reproduction through male killing, feminization, parthenogenesis, and cytoplasmic incompatibility [2, 5]. In this regard, manipulation of host reproduction by Wolbachia is an important consideration in plant-insect systems and often plays an important role in a variety of evolutionary processes, including local adaptation, gene flow, host-associated differentiation, and speciation [6, 7].
In particular, Wolbachia induced cytoplasmic incompatibility (CI) has important evolutionary implications for insect populations distributed across space and differing ecological environments. Cytoplasmic incompatibility results in male and female gametes being unable to form viable offspring due to differences in parental Wolbachia infection status. When Wolbachia infected males mate with uninfected females (unidirectional infection) few or no offspring are produced while all other crosses are fertile [8]. As a result, once infection levels in a population surpass a threshold, Wolbachia is predicted to sweep through the host population [9,10,11]. The reproductive advantage of Wolbachia infected individuals can result in rapid spread of the endosymbiont [9, 12]. Moreover, the co-maternally inherited mitochondrial DNA can hitchhike with the spreading Wolbachia replacing mitochondrial haplotypes associated with uninfected individuals [11, 13]. Thus, Wolbachia infected populations typically exhibit lower mitochondrial diversity than uninfected populations [13,14,15]. Wolbachia infected populations are also usually associated with specific mitochondrial haplotypes i.e., those of their founder individuals which acquired Wolbachia horizontally from other hosts [12] or due to hybrid introgression [16].
When both partners are infected with different strains of Wolbachia, bidirectional CI occurs with both mating directions being infertile. This has peaked interest in the possible role of Wolbachia in promoting speciation or maintaining species boundaries [6, 7, 17, 18]. In essence, bidirectional CI can increase reproductive isolation by decreasing gene flow among populations that harbor different Wolbachia strains [19]. The presence of CI can also produce selection for other pre-zygotic isolating mechanisms. For example, Wolbachia induces behavioral isolation in mushroom feeding Drosophila via selection against heterospecific incompatible matings of uninfected females with infected males [20]. Therefore, the endosymbiont may promote divergence even within species that are infected by only one Wolbachia strain. Further, different Wolbachia strains promote pre- and postzygotic isolation in the Drosophila paulistorum species complex where the symbiont acts as a pathogen in hybrid males causing both embryonic inviability and male sterility [21]. Bidirectional CI is the primary form of reproductive isolation between species of Nasonia wasps: Nasonia giraulti and Nasonia longicornis harbor different Wolbachia strains and are reproductively isolated [19].
Given the wide-ranging effects of Wolbachia infection in generating and/or maintaining barriers to gene flow among populations, and our prior knowledge of the ecology and biology of the gall-forming wasp, Belonocnema treatae, Mayr (Hymenoptera: Cynipini: Cynipidae) we surveyed the infection status of host-associated populations of this cynipid. This species is a regional host specialist on closely related live oak species (genus Quercus; subsection Virentes) that are distributed across the southeastern U.S.: Q. fusiformis (Qf), Q. virginiana (Qv) and Q. geminata (Qg) (Fig. 1). Populations of B. treatae on Qv and Qg exhibit both partial positive assortative mating and host-associated habitat preference, and express significant differences in adult and gall morphology [22,23,24] in areas where the two host plants overlap. However, pairwise comparisons of neutral mtDNA markers among B. treatae populations resident on Qv and Qg across Florida revealed that sequence divergence is not associated with host use, as allopatric populations on the same host exhibited similar patterns of divergence compared to allopatric populations on different hosts [22]. Thus, these populations of B. treatae appear to be at the incipient stage of adaptive differentiation and speciation. What is currently not known is whether Wolbachia infection and/or strain diversity is involved in the partial reproductive isolation that is observed between different host-associated populations of B. treatae and whether Wolbachia is associated with potential population genetic structure of B. treatae evident on larger scales (i.e., across the geographic range of B. treatae in the southern U.S.).
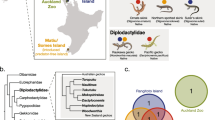
Wolbachia infection frequencies in each of 23 Belonocnema treatae populations distributed across the southern United States and sampled from three host plant species. Filled circles = Quercus fusiformis, filled squares = Quercus virginiana and filled triangles = Quercus geminata. Pie charts represent the percentage of B. treatae infected by Wolbachia in each population: white = uninfected individuals, yellow = wTre1, blue = wTre2, red = wTre3, grey = wTre4, orange = wTre1&3, green = wTre2&3, and purple = wTre1&2&3. Sample size per population is denoted by size of the pie diagram. Details of population localities, sample size, sample composition and infection frequencies are given in Table 1. Map adapted from Wikimedia commons (CC BY-SA 3.0)
The life cycle of B. treatae includes temporally and spatially isolated sexual and asexual generations (i.e., cyclical parthenogenesis or heterogony), common among cynipid gall wasps [25]. Wolbachia’s role as a sex ratio distorter [2] suggests a potential contribution of this endosymbiont in life cycle diversity of B. treatae. For example, Plantard et al. [26] demonstrated geographic variation in the prevalence of Wolbachia infection in the rose gall wasp Diplolepsis spinosissimae and showed that the endosymbiont is associated with obligate homozygous parthenogenesis. However, geographic variation in the prevalence of Wolbachia infection, infection with phylogenetic different Wolbachia strains, and the occasional occurrence of males highlights the complex infection history in this gall wasp species [26, 27]. Wachi et al. [28] concluded that Wolbachia induced CI may have led to the evolution of reproductive isolation between closely related oak gall wasps and thus contributed to the evolution of its host. In contrast, Abe et al. [29] described the occurrence of Wolbachia in the Andricus mukaigawae complex in Japan but did not detect an association of Wolbachia in bivoltine heterogonic and univoltine thelytokous species, thus excluding a potential role of the endosymbiont in the life cycle of this species complex. Screening of 64 gall wasp species by Rokas et al. [30] showed that Wolbachia is rare in oak gall wasps with just five species infected: three cyclical parthenogens (exhibiting both sexual and asexual generations) and two thelytokous species (exhibiting only the asexual generation). The authors excluded Wolbachia as a causative agent of life cycle diversity within the Cynipidae [30]. Although, this study found variable infection rates among species and populations within species, in some cases the limited number of samples examined (single individuals sampled per population, or single populations per species) may underestimate the number of species infected by the endosymbiont and thus might have underestimated the potential role of Wolbachia in the evolution of cynipids.
Our goal is to understand the ecological and evolutionary patterns and consequences of Wolbachia infection within and among populations of B. treatae. Herein, we address seven questions: (1) Given the large geographic range of B. treatae, we examine whether geographically separated populations within each host plant species display variable infection rates by the same or different Wolbachia strains? (2) Given that host-associated B. treatae populations exhibit partial reproductive isolation [22, 23], we ask whether populations associated with alternative host plant species differ in infection status or harbor different Wolbachia strains? (3) Given the generational differences in reproductive mode (parthenogenetic versus sexual reproduction), the presence of haploid males and diploid sexual females, development on different plant parts and at different times, and dramatic differences in the diversity and mortality attributed to insect parasitoids and inquilines [31, 32], we ask whether infection status differs between generations and between sexual males and females? (4) Given the prediction of reduced mitochondrial diversity due to the potential for selective sweeps of Wolbachia within infected populations, we inspect the relationship between Wolbachia infection and mitochondrial diversity of its host. (5) From the perspective of potential coevolution with the host, we ask whether the maternally inherited endosymbiont is associated with specific mitochondrial haplotypes of the host? (6) We ask, what is the phylogenetic relationship among Wolbachia strains resident in B. treatae? If closely related Wolbachia strains that are detected within B. treatae form a clade, this would suggest a deeper long-term association of the symbiont with B. treatae. In contrast, if multiple unrelated Wolbachia strains are present and distributed through the tree, repeated acquisition of Wolbachia by different gall wasp populations is implicated. (7) Finally, to further understand sources of among population variation in infection rates, we address the vertical transmission rate of a Wolbachia strain. Incomplete maternal transmission rates might explain different infection frequencies and potentially occasional loss of the endosymbiont.
To address these questions, we (1) sampled asexual and/or sexual generations of B. treatae from 23 populations distributed across a ~ 2400 km transect that spans the southern U.S. from all three host plants, (2) screened individuals for Wolbachia infection and examined strain diversity by sequencing a fragment of the Wolbachia surface protein (wsp) gene of all infected individuals, (3) sequenced a fragment of the mitochondrial cytochrome oxidase I gene of B. treatae to construct a phylogeny of B. treatae populations and (4) screened parents and offspring from single pair crosses to estimate vertical transmission rates of Wolbachia. We then mapped Wolbachia infection status and strain diversity across generation, geography, host plant affiliation and B. treatae phylogeography. Collectively, our results highlight a long evolutionary history of Wolbachia in B. treatae with complex infection dynamics across the geography, ecology, and life history of its host.
Methods
Study system
Gall wasps, family Cynipidae, comprise > 1300 species of secondarily phytophagous parasitic Hymenoptera [33,34,35]. Many cynipids, including B. treatae [36], exhibit heterogony (cyclical parthenogenesis), in which temporally and spatially segregated sexual and asexual generations alternate to complete the life cycle [25, 37]. The three species of host plants used by B. treatae in the southern U.S. exhibit partially overlapping geographic distributions (Fig. 1; [24, 38, 39]). Sexual generation B. treatae develop within multi-chambered root galls during the winter and emerge in the spring. Following mating, sexual females oviposit into the undersides of new leaves distributed throughout tree crowns and induce galls that each house a single asexual gall wasp. Asexual generation B. treatae consist of androphores and gynophores, which typically produce all male or all female sexual generation broods within root galls. Each root gall typically contains siblings produced by a single asexual female. Therefore, when surveying for Wolbachia (see below) within the sexual generation we screened only one individual from each root gall to avoid biased estimates of infection. The two generations do not overlap in time, exploit very different environments and are exposed to differing natural enemy communities. Asexual generation B. treatae developing in leaf galls in the crowns of trees are attacked by at least 20 species of insect natural enemies (parasitoids and inquilines) while the underground sexual generation developing in root galls are attacked by only 4 species [32]. Moreover, the percent of asexual leaf galls in which parasitoids and inquilines develop is far greater than in root galls housing the sexual generation. These spatial and temporal differences in the timing of galler development [31] and dramatic differences in natural enemy pressure [32] are possible sources of differential acquisition of Wolbachia between the generations and motivate the inspection of Wolbachia infection and strain diversity within the life cycle of B. treatae.
Specimen sampling
We sampled 23 B. treatae populations distributed across the range of the three host plants (five Qf associated populations, ten Qv populations, and eight Qg populations) in Oklahoma, Texas, Mississippi, Alabama, Florida, and Georgia (Fig. 1; Table 1). Asexual adults were obtained from 16 populations by haphazardly collecting between several hundred to several thousand leaf galls from multiple locations within the crowns of trees at each site. Leaf galls were then binned by site in bulk collection traps. Sexual adults were obtained from 15 populations by haphazardly collecting root galls excavated from beneath heavily leaf-galled trees from each site. Root galls were returned to the laboratory and housed individually in standard fruit fly rearing vials. Galls of both generations were maintained outdoors at the Texas State Greenhouse in a shaded alcove under natural conditions. Collections were monitored daily and upon emergence, live adults were immediately stored in 95% ethanol. Details of population locations, host plant affiliation, generation(s) sampled, and sample sizes are shown in Table 1. Genomic DNA was then extracted from whole adult bodies using DNeasy Blood and Tissue kits (Qiagen Inc., Valencia, CA).
Wolbachia genotyping
A total of 463 B. treatae individuals from the 23 populations were screened for Wolbachia using the primers 81F and 691R [40] that target a ~ 600 bp fragment of the Wolbachia surface protein wsp gene. We performed PCR reactions in a total volume of 10 μl using 10× reaction buffer Y (Peqlab, Germany), 800 μM dNTPs, 0.3 μM of each Primer, 0.5 U peqGold Taq DNA polymerase (Peqlab, Germany) and 1 μl of template DNA. All reactions were performed on a 2720 thermal cycler (Applied Biosystems) with the following conditions: 94 °C for 4 min, followed by 34 cycles at 94 °C for 30 s, 55 °C for 1 min, 72 °C for 1 min and a final extension at 72 °C for 10 min. All PCR products were run on a 2% electrophorese gel and visualized by Gel Red nucleic acid gel staining and UV illumination. To exclude false positives and/or negatives, all PCRs were replicated twice to confirm results. All positive samples were then Sanger sequenced by Eurofins MWG Operon (Ebersberg, Germany). Sequences were individually assembled and manually edited using CodonCode Aligner vers. 3.7.1 (CodonCode Corp., Centerville, MA). All sequences were then screened for ambiguous sites by inspecting all raw chromatograms for multiple peaks, to determine if individuals were infected by multiple Wolbachia strains.
To segregate alleles of potentially multi-infected individuals we cloned PCR products from nine individuals selected from geographically separated populations that either showed double and or triple peaks indicating the presence of multiple Wolbachia strains. Additionally to check for potentially hidden Wolbachia strains (e.g. [41]) we cloned eight individuals whose chromatograms showed clear single peaks suggesting the presence of just one Wolbachia strain. Molecular cloning was performed by Eurofins MWG Operon (Ebersberg, Germany) using pTZ57R/T vector (Thermo Scientific, USA) and a TOPO TA cloning kit. Eleven to 19 plasmids per individual were Sanger sequenced and the 286 sequences were screened and edited manually and aligned using CodonCode Aligner.
Mitochondrial genotyping
We further genotyped a 633 bp region of the mitochondrial cytochrome oxidase I (COI) gene of all 463 B. treatae individuals. Individuals were sequenced using the barcode primers LCO 1490 and HCO 2198 [42]. PCR reactions were performed in a total volume of 10 μl containing 10× reaction buffer Y (Peqlab, Germany), 800 μM dNTPs, 0.3 μM of each Primer, 0.5 U peqGold Taq DNA polymerase (Peqlab, Germany) and 1 μl template DNA. The thermocycler protocol was: 94 °C for 3 min, followed by 34 cycles at 94 °C for 30 s, 55 °C for 45 s, 72 °C for 45 s and a final extension at 72 °C for 7 min. All PCR products were run on a 2% electrophorese gel, visualized by Gel Red nucleic acid gel staining and then Sanger Sequenced by Eurofins MWG Operon (Ebersberg, Germany). Resulting sequences were edited manually and aligned using CodonCode Aligner.
Genetic distance and nucleotide diversity within and among B. treatae populations
To characterize patterns of genetic variation within and among B. treatae populations, we calculated the average genetic distance between and within groups of B. treatae (i.e., host plant-affiliated populations, population pairs, between generations and sexes), measured by the number of nucleotide substitutions occurring between two mtDNA haplotypes using the Tamura Nei model [43] determined by the best fitting model option in MEGA. All analyses were calculated in MEGA v. 6.06 [44]. Given the phylogenetic results (see below), COI haplotypes were then grouped for additional comparisons into western (1–8) and eastern populations (10–23) according to their level of sequence divergence and Wolbachia infection status. Population 9 (Gautier, Mississippi) was excluded from this specific analysis as it contained a mix of eastern and western B. treatae haplotypes and Wolbachia strain infections.
Phylogenetic history of Wolbachia and B. treatae
To infer the evolutionary history of Wolbachia infection in B. treatae and to assess whether Wolbachia influenced the mitochondrial diversity of its host, we performed a phylogenetic analysis of B. treatae populations using the mtDNA sequence data. We followed the phylogenetic methods of Ronquist et al. [45] that performed a similar analysis using the COI gene for > 100 cynipid gall wasp species, including several taxa from North America closely related to the genus Belonocnema. All individuals from each of the 23 populations were included. Our analysis contained four cynipid outgroup taxa: Biorhiza pallida (GenBank accession number: AY368931), Cynips quercus (DQ012638), Trigonaspis mendesi (DQ012658) and Neuroterus numismalis (AY368930) previously published in [45]. Phylogenetic relationships were estimated using Bayesian inference implemented in MrBayes version 3.2.2 [46] following the methods optimized for mtDNA COI analysis of gall formers detailed in [45]. Here, sequence data was modeled using the four by four nucleotide model that estimates stationary state frequencies that integrate across all grouping and ungrouping exchangeability rate parameters [45, 47]. In addition, we used the mixed-rates evolutionary model with a gamma distribution which allows substitution rates to vary under a flat Dirichlet (default) prior. The protein-coding COI gene was divided into two categories, first and second codon positions and third positions, with model parameters and base rate substitutions uncoupled [45, 46]. Default setting was used for all remaining parameters. The model was run four independent times using four Metropolis-coupled chains, interactively, for three-million generations at a time, until the deviation of split frequencies converged on a value < 0.01 (~ 21 million generations). Chains were sampled once every 1000 generations with an initial burn in of 25%. In each analysis, all branch lengths and substitution model parameters possessed potential scale reduction factors between 1.00 and < 1.01. We then constructed a majority rule consensus tree (presented herein) from the post burn in data.
To provide insight into whether Wolbachia infections of B. treatae populations observed in our study more likely represent deep ancestral associations with the symbiont, or repeated acquisitions of Wolbachia by different gall wasp populations we compared the phylogenetic relationships of the four Wolbachia strains found in B. treatae using the wsp gene sequences with representative Wolbachia sequences from other insect systems. To form the phylogeny, we used a consensus wsp sequence for each of the three major Wolbachia strains (wTre1–3) found in B. treatae, sequences from related Wolbachia strains available from GenBank and included the rare B-group strain wTre4 as an outgroup. Similar to the methods employed for B. treatae, we used MrBayes version 3.2.2 [46] to construct the phylogeny with the following exception: we used the GTR + I + G model of sequence evolution adopted from [48] which was run four independent times for 50,000,000 generations until the deviation of split frequencies converged on a value of 0.003.
Vertical transmission rate of Wolbachia
A concurrent study of multiple paternity in B. treatae from single pair crosses involving an additional 353 individuals allowed us to examine the vertical transmission rate of Wolbachia from adult sexual generation females to their asexual offspring. This study involved the San Marcos, Texas population (population 3) that was characterized by intermediate infection frequencies of the endosymbiont. Infected individuals carried strain wTre1 (see results below). We assayed the Wolbachia infection status of 23 sexual generation females and eleven to 16 asexual progeny produced by each female. The frequency of infection of asexuals produced by infected sexual females estimates the vertical transmission rate of the wTre1 Wolbachia strain during this phase of the B. treatae life cycle.
Statistical analysis
We tested for heterogeneity in the frequency of infection among B. treatae populations and the influence of host plant association, geography and cynipid generation on the average frequency of Wolbachia infection. To test for differences in Wolbachia infection among populations we used χ2 tests. We first tested for variation among all populations and then subsetting the data we tested for variation among populations within alternative generations (asexual versus sexual), geography/phylogenetic clades (east versus west clade), and host plant association (Qv, Qg, Qf). Given multiple testing, we adjusted our alpha value by the number of tests run from the same dataset to control for Type 1 error (α = 0.05/4 = 0.0125). To test the influence of host plant association and generation on average infection frequency, we ran a t-test or ANOVA on arcsine square root transformed population infection frequencies weighted by the sample size. All statistical analyses were performed in R vers. 3.3.1 [49].
Results
Wolbachia infections dynamics within and among B. treatae populations
Across all B. treatae populations, 56.1% of the 463 individuals screened were infected by Wolbachia. Infection frequency varied markedly among the 23 populations (X2 = 409, df = 22, P < 0.0001; Fig. 1). We found 100% infection in ten populations and 0% infection in six populations (Table 1). In seven populations, both infected and uninfected individuals were present (Table 1), with infection frequencies ranging from 11% to 95%. When B. treatae populations were grouped into western and eastern clades based on mtDNA haplotypes (see below), the Wolbachia infection frequency differed among populations within both the western clade (X2 = 190, df = 7, P < 0.0001) and the eastern clade (X2 = 190.57, df = 13, P < 0.0001). There was no association between the Wolbachia infection status (infected or uninfected) and mtDNA haplotype (X2 = 0.01, df = 91, P = 0.9591; Fig. 2).
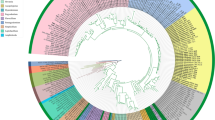
Bayesian phylogenetic tree based on sequencing of the mtDNA COI gene for 463 Belonocnema treatae sampled from 23 populations. Branch colors represent geographic separation between populations from West (light blue; populations 1–8) and East (orange; populations 10–23) and population 9 (grey). The outside bar represents the host plant affiliation for each collection site (red = Q. fusiformis, green = Q. geminata and blue = Q. virginiana). The different dots represent the infection status of each single individual with white = uninfected, yellow = wTre1, blue = wTre2, red = wTre3, grey = wTre4. Individuals with two or three dots indicate infection by more than one Wolbachia strain
We also tested for variation in Wolbachia infection frequency across B. treatae populations affiliated with each of the three host plant species. We found Wolbachia infection frequency to be 37.5% for individuals sampled from Qg (N = 136), 61.1% for individuals sampled from Qv (N = 162), and 66.7% for individuals from Qf (N = 165). However, within each host plant association, infection frequencies among populations were highly variable (Qg: X2 = 129.1, df = 7, P < 0.0001; Qf: X2 = 149.8, df = 4 P < 0.0001; Qv: X2 = 127.6, df = 9, P < 0.0001). ANOVA of population infection frequencies weighted by sample size showed no evidence of statistically significant differences in infection frequencies among the three host plants (F2,20 = 0.54, P = 0.5910). Notably, we found populations with 100%, 0%, and partial infection frequencies associated with all three host plants.
Comparison of the frequency of Wolbachia infection between the generations across all populations showed no general difference between asexual (0.53 ± 0.11 SE) and sexual (0.62 ± 0.09 SE) individuals (t = 0.75, df = 29, P = 0.4574). Moreover, the eight sites containing samples from both sexual and asexual generations provided a direct comparison of infection rates among the alternate generations while excluding the effects of variation in infection frequencies among populations (Additional file 1: Table S1). This direct comparison demonstrated near identical Wolbachia infection frequencies between the sexual and asexual generations of B. treatae (paired t-test: t = 1.23, df = 7, P = 0.2583).
High mitochondrial diversity within B. treatae
Characterization of the 633 bp fragment of the mtDNA COI gene of 463 B. treatae individuals sampled from 23 populations distributed across the southern U.S. yielded 92 distinct haplotypes (GenBank Accession Numbers MG252379–252470). Haplotypes clustered into two major clades: a western clade encompassing populations from Oklahoma, Texas and Mississippi (populations 1–8) and an eastern clade encompassing populations from Florida, Georgia and Alabama (populations 10–23) (Fig. 2). Population 9, from Gautier, Mississippi, geographically located between populations grouping with the eastern and western clades, contained individuals with haplotypes from both the eastern and the western clades (Figs. 1 and 2).
Sequence divergence between individual mtDNA haplotypes ranged from 0.2% to 7.2% indicating considerable variation in the degree of divergence among the 23 B. treatae populations. The genetic distance between western and eastern clades was 6.3%, while the mean genetic distance between pairs of populations that clustered within the western and eastern clades was 0.5% and 2.1%, respectively (Table 2). Within each of the three sets of host plant-affiliated populations the mean genetic distance between pairs of populations was 0.4% in Qf, 3.8% in Qv and 1.6% in Qg indicating greater among population genetic distance among populations resident on Qv. When the ten B. treatae populations that used Qv as the host plant were binned into eastern and western clades, the mean genetic distance between pairs of western populations of B. treatae on Qv was 0.4% compared to 2.2% between pairs of eastern populations (Table 2). Finally, the average pairwise genetic distance among sexual (0.4%) and asexual (0.5%) populations in the western clade was lower than corresponding observed average genetic distances in the eastern clade (2.1% sexual generation and 2% in the asexual generation). These generation-specific estimates of genetic distance among populations accord with the average genetic distance estimates among pairs of populations clustering in the western and eastern clades (above) and suggest no influence of asexual versus sexual reproduction on sequence divergence (Table 2).
Wolbachia strain diversity and co-occurrence in B treatae populations
By sequencing the wsp gene of all 260 Wolbachia infected individuals and 286 clones derived from 17 individuals, we found that B. treatae is infected by four Wolbachia strains designated wTre1–4 (GenBank accession numbers MG252471–252474). Strain wTre1 was mainly present in the western B. treatae mitochondrial clade, which included populations from Oklahoma, Texas and western Mississippi. In this clade, all infected individuals harbored only wTre1, with the exception of a single individual sampled in Luling, Texas, confirmed by TOPO cloning to be double infected by wTre1&3 (Fig. 1). The three other Wolbachia strains were found exclusively in the eastern B. treatae clade. Strain wTre2 was present in varying frequencies in populations sampled in Mississippi, Alabama and northern Florida while individuals from Gautier, Mississippi (population 9), were either infected by wTre1 or by wTre2. Strain wTre3 was present in two populations in northern Florida and one population in southern Florida, while wTre4 was detected in only two individuals from Perry, Florida (population 15). All B. treatae individuals from Sapelo Island, Georgia (population 16) were confirmed to be double infected by wTre2&3 and all but one individual collected at Hickory Hammock, Florida (population 21) and Kissimmee River, Florida (population 22) was triple infected by wTre1&2&3, with a single individual found to harbor strains wTre1 and wTre3 only (Fig. 1).
The phylogeny of the wsp gene shows that the three major Wolbachia strains (wTre1–3) detected in B. treatae belong to the Wolbachia supergroup A. Strain wTre4, however, belongs to the supergroup B (Fig. 3). Within the A-group, the three B. treatae associated strains were more closely related to Wolbachia strains from other species and geographic localities than they were to each other (Fig. 3). This pattern is consistent with B. treatae acquiring multiple infections through time.
Bayesian phylogenetic tree based on wsp surface protein gene sequences for the three most common Wolbachia strains wTre1, wTre2 and wTre3 from supergroups A and a sample of related Wolbachia sequences from insect taxa obtained from GenBank. We used the rare B-group strain wTre4 as outgroup. Bootstrap values above 50% are shown. The branch between supergroup A and supergroup B was reduced as indicated by double-bars
Wolbachia and haplotype diversity of B. treatae
There was no strict association between B. treatae mtDNA haplotype and Wolbachia infection status across the range of the insect host (Fig. 2). However, inspection of B. treatae populations that contained both infected and uninfected individuals or individuals infected by different Wolbachia strains suggest effects of the endosymbiont on haplotype diversity of its host. For example, mitochondrial haplotypes that clustered either into the western or eastern clade were associated with their Wolbachia infection. Specifically, wTre1 infected individuals were exclusively associated with western haplotypes and wTre2 infected individuals were exclusively associated with eastern haplotypes (Fig. 2). Although there was no strict range-wide association of Wolbachia strain and mtDNA haplotype, two of the 23 populations did exhibit such patterns: Individuals from Gautier, Mississippi (population 9), were either infected by wTre1 or by wTre2, where wTre1 was associated directly with a western mtDNA haplotype and wTre2 was associated with an eastern mtDNA haplotype. We found a similar pattern in Cedar Key, Florida (population 19), where individuals were either infected by wTre2 or wTre3. The mtDNA sequence diversity within wTre2 infected hosts was 0.3% while all wTre3 infected gall wasps shared the same haplotype. In contrast, the sequence divergence between individuals infected by the two different Wolbachia strains was 2.6% which is consistent with a strict association of Wolbachia with specific mitochondrial haplotypes at the population level. In the population from Progress Park, Florida (population 18), we found individuals that were uninfected, one individual infected by wTre2 and individuals infected by wTre3 (Fig. 1). While the wTre2 infected and uninfected individuals shared the same haplotype, the wTre3 infected gall wasps exhibited different mtDNA haplotypes with a sequence divergence of 0.5%.
The strain wTre1 was present in B. treatae developing on both plant hosts, Qf and Qv, in the west. Similarly, wTre2 and wTre3 were detected in B. treatae developing on Qg and Qv in the east. The double and triple infection in the eastern clade, however, occurred exclusively on individuals attacking Qv. The three strains occurred in both asexual and sexual B. treatae generations while wTre4 was detected exclusively in two sexual females from Perry, Florida (population 15).
In addition to identifying the four Wolbachia strains, sequencing of 286 clones from 17 individuals indicated the presence of widely dispersed SNPs across the 600 base pair fragment of the wsp gene. High SNP diversity was found in wTre1 (100 SNPs in 126 clones), wTre2 (54 SNPs in 67 clones) and wTre3 (78 SNPs in 93 clones). Of the total of 232 SNPs detected, 96% were singletons and just ten SNPs were confirmed by independent PCRs. Although we cannot rule out occasional sequencing and cloning errors, the high incidence of SNPs detected within strains suggests that it is likely that different subtypes of the four main strains are present.
Wolbachia transmission frequency between generations
Of the 23 sexual females examined to assess transmission of Wolbachia to their asexual progeny, ten females were positive for Wolbachia while 13 were not infected by the endosymbiont. Within infected females in three cases, 100% of the offspring (N = 14–16 per female) were also infected by the endosymbiont suggesting perfect maternal transmission (Additional file 2: Table S2). However, in seven cases, Wolbachia transmission was imperfect with transmission rates ranging from 64% to 94%. The average transmission rate estimated across the ten females was 87.5% (Additional file 2: Table S2). Further, 182 progeny (N = 14 per female) of the 13 females not infected by Wolbachia were additionally tested. For all 13 females, 100% of their offspring was also tested negative (Additional file 2: Table S2).
Discussion
We examined a widely distributed North American species of cynipid gall forming wasp that exhibits regional host specialization and asked whether patterns of Wolbachia infection and diversity are associated with the insect’s host plant association, geographic distribution, mitochondrial evolutionary history, and life cycle. By analyzing Wolbachia infection status and strain diversity within and among individual gall formers drawn from 23 B. treatae populations distributed across the three host plant species sampled from across the southern U.S., we demonstrated highly variable Wolbachia infection and high strain diversity among populations. We found little evidence that host plant affiliation of the gall former was associated with Wolbachia infection status, however we did find evidence of an association between Wolbachia strain and geography of the host. Finally, we detected Wolbachia in equal frequencies in both asexual and sexual generations and between males and females of the sexual generation. This conforms with previous studies that suggest that Wolbachia may not influence the sex determination system in gall wasps [26, 29, 30].
Diversity and distribution of Wolbachia in divergent B. treatae lineages
In contrast to patterns predicted by selective sweeps of mtDNA haplotypes associated with Wolbachia induced CI [11, 15, 50, 51], we detected no difference in mitochondrial haplotype diversity between Wolbachia infected and uninfected populations of B. treatae. However, we did find strong associations between Wolbachia strain type and the geographic distribution of B. treatae: strain wTre1 is almost exclusively present in populations from the western portion of the geographic range of B. treatae, while strains wTre2 and wTre3 are almost exclusively present in populations from the east. Strain wTre4 was a low frequency strain found only in two individuals in one population from Florida (Fig. 1). This pattern of distribution of Wolbachia strain types was matched by the significant break in the mtDNA gene tree between eastern and western B. treatae populations (Fig. 2).
Two alternative scenarios could explain the observed distribution patterns of the three most frequent strains, wTre1–3. First, Wolbachia infection in B. treatae may be an ancient infection present before the divergence of B. treatae into eastern and western mtDNA clades. In this scenario, the three major Wolbachia strains have evolved following the geographic separation of the host. While, secondary endosymbionts like Wolbachia are able to cross species boundaries and can therefore be randomly distributed among different hosts [52, 53] coevolution is also possible where Wolbachia can persist in host lineages more than a million years [54]. Alternatively, B. treatae populations may have acquired Wolbachia strains independently after the divergence of eastern and western mtDNA clades. The fact that different wTre strains do not form a monophyletic clade but rather are distributed across the Wolbachia phylogenetic tree and are related to strains found in other insect hosts (Fig. 3), suggests that B. treatae populations have independently acquired different Wolbachia strains after separation into eastern and western clades. This scenario also conforms with the observation that distinct eastern and western mitochondrial haplotypes are co-associated with the same Wolbachia strain, further suggesting that independent horizontal transmissions of Wolbachia could have produced this pattern. Under this scenario B. treatae populations that are not infected by Wolbachia simply reflect populations that have not yet been invaded by the endosymbiont. However, as supported by our findings of incomplete vertical transmission of the symbiont, these uninfected populations may also have lost their associated wTre strains. Within well studied populations of live oaks a minority of individual live oak trees that are patchily distributed typically support the vast majority of B. treatae [55]. Further these infrequent trees harboring B. treatae experience orders-of-magnitude among-year-variation in the abundance of B. treatae, (personal observation). Together, these features of the ecology of B. treatae in concert with incomplete vertical transmission may set the stage for periodic loss of infections.
To resolve if a Wolbachia driven selective sweep has influenced the mitochondrial structure of its host, future studies need to examine nuclear genetic markers. While a disconcordant pattern of mitochondrial and nuclear DNA would support the hypothesis of a Wolbachia-driven selective sweep, a concordant pattern would suggest that other factors such as the demographic history of the host have driven the observed mitochondrial divergence [56]. Moreover the microbial community of B. treatae needs to be characterized more comprehensively to understand if any other symbiont might have influenced the observed pattern.
B. treatae populations harbor multiple Wolbachia strains
Two intriguing aspects of our results are the detection of multiple Wolbachia strains within five of the 23 surveyed B. treatae populations and the presence of multiple strains within individual gall formers. Study-wide, among the 260 individuals infected with Wolbachia, we found that 86% of individuals harbored a single Wolbachia strain. In population 9 in Gautier, Mississippi, we found individual B. treatae that were infected with wTre1 or wTre2 but none of the 28 individuals examined were infected by both strains. Similarly, individuals from populations from Cedar Key, Florida (population 19), and Progress Park, Florida (population 18), were infected by either wTre2 or wTre3 but no individual in these populations were simultaneously infected by both strains. In contrast, these same two strains occur as a double infection in all ten individuals sampled from Sapelo Island, Georgia (population 16), and all three major wTre strains occur within the same individuals from central Florida (Hickory Hammock [population 21] and Kissimmee River [population 22]; Fig. 1; Table 1).
The presence of multiple Wolbachia strains within single individuals has been documented in several species [41, 57,58,59] including three gall wasp species [30]. In such cases the horizontal acquisition of a new strain [12] or a spreading infection from a restricted host population [11] can cause host populations to acquire additional strains. Other cases, however, suggest that a spreading new Wolbachia strain is displacing a previously present infection [10]. Lastly, theoretical models have shown that spatial structure and habitat fragmentation can promote the coexistence of Wolbachia [60], however the general mechanisms whereby individual insects acquire and maintain multiple strains are not clearly understood and requires further work.
Most of the double and all of the triple infected individuals that we detected clustered together on one branch of the mtDNA tree (Fig. 2) along with some uninfected and single infected individuals. All of the single infected individuals on this branch are infected by wTre2 suggesting that the double infections present in all individuals in Sapelo Island, Georgia (population 16) might have resulted from a horizontal acquisition of wTre3 by previously wTre2 single-infected individuals. Similarly, in the triple infected populations from central Florida, previously wTre2 infected individuals might have acquired both wTre1 and wTre3 horizontally. It is unclear why the wTre1 infection that occurs in multiple western populations as the only strain, appears as a triple infection in central Florida.
Parasitoids and inquilines are possible sources of horizontal Wolbachia transmission [61,62,63]. Belonocnema treatae is attacked by a diverse natural enemy community composed of parasitoids and inquilines [32], of which some species are known to harbor Wolbachia infections e.g. [64, 65]. This natural enemy community represents an extensive source of potential transmitters of Wolbachia strains. More broadly the three species of live oaks that are galled by B. treatea are also the host plants of a diverse community of cynipid gall formers. The shared habitat of communities of gallers and their associated natural enemies [24, 66, 67] represents a complementary avenue of direct or indirect Wolbachia exchange. Thus, a possible explanation for the occurrence of wTre1 in the eastern clade is horizontal transmission of Wolbachia by parasitoids, inquilines, or other gall wasp species co-occurring on the same host plants. Studies of communities of gall wasp species and their respective communities of parasitoids and inquilines associated with shared host plant species e.g. [57] are needed to elucidate the transmission and subsequent population dynamics of single and multiple strains of Wolbachia within and among trophic levels for these insect communities. Additionally, studies that examine the relationship between variation in parasitoid pressure (i.e., rate of parasitism and diversity of parasitoids) among B. treatae populations with different Wolbachia infections are needed to understand the distribution of Wolbachia in B. treatae.
An alternative explanation of the occurrence of wTre1 in eastern populations (Fig. 1) is that the eastern and the western wTre1 infections actually constitute different Wolbachia strains that share the same wsp alleles and hence are not distinguishable by our study. Additional multilocus sequence typing (MLST) of five conservative genes can resolve hidden genetic diversity of Wolbachia [68]. To gain more insight into the evolutionary history of wTre strains in B. treatae a future study will focus on a more extensive genomic characterization of the four wTre strains detected in this host species to resolve potential co-evolution of the endosymbiont with its host. However, delineation of closely related strains can still be challenging when using MLST [69, 70]. Thus, in addition to characterizing additional MLST genes, a comparative genomic approach involving sequencing of individual B. treatae and Wolbachia from diverse populations will aid in confirming both the relationships among the putatively different wTre strains and delineating population substructure of the host.
Potential role of Wolbachia in reproductive isolation of B. treatae
The potential role of Wolbachia as a driving factor in insect speciation remains controversial [2, 6, 7, 18]. Wolbachia infection needs to fulfill two conditions to cause speciation: 1) infection polymorphisms among different host populations must be stable and 2) cytoplasmic incompatibility must be strong enough to generate natural selection favoring the evolution of reproductive isolation which promotes divergence of its host [2]. Unidirectional CI appears insufficient as it results in unstable equilibrium frequencies and therefore the spread of the bacterium is predicted to occur more rapidly than the evolution of reproductive isolation [9, 20]. Importantly however, theoretical predictions show that two bidirectional CI-inducing Wolbachia strains can be sufficiently stable and demonstrate that gene flow can be substantially reduced between hosts harboring different strains [71, 72]. However, few studies have demonstrated coexistence of different Wolbachia strains along a contact zone [73]. In our study, while most infected B. treatae populations are infected by a single Wolbachia strain we also found three populations containing individuals multiply-infected by different wTre strains. The occurrence of different Wolbachia strains within B. treatae populations highlights the potential of Wolbachia to contribute to the diversification in this gall wasp. In particular, the strict association of Wolbachia with the mitochondrial haplotype of the individuals in Gautier, Mississippi, and Cedar Key, Florida, where the Wolbachia infection status of all 39 individuals corresponds to a specific haplotype, further suggests a potential role of Wolbachia in reproductive isolation with either no hybridization between gall wasps infected by different wTre strains or perfect CI with complete maternal transmission of Wolbachia. A preliminary analysis of population genetic structure of B. treatae throughout its geographic range based on over 41,000 SNP loci supports the presence of two independent B. treatae lineages in the Gautier population (unpublished data). Future studies of the dynamics of Wolbachia strains in B. treatae should include characterizing the phenotypic effects of the different wTre strains, further exploration of populations with two or more strains, and detailed crossing studies to address CI directly. If these strains are shown to cause CI, the co-occurrence of individuals infected by different strains could lead to bidirectional CI. In this case Wolbachia could act as a potential factor in driving reproductive isolation of its host e.g. [19]. Detailed inspection of the populations in Gautier, Mississippi, where wTre1 and wTre2 infected B. treatae individuals meet in a population that may represent a zone of secondary contact, and populations in Cedar Key, Florida, where wTre2 and wTre3 infected individuals co-occur, will show to what degree the two strains might cause CI and influence pre- and postzygotic isolation in B. treatae [22, 23]. Finally, crossing studies should investigate the maternal transmission efficiencies of the four Wolbachia strains as a means of explaining the absence of the endosymbionts in populations containing uninfected individuals.
Conclusion
In this study, we surveyed for the presence and diversity of Wolbachia in the heterogonic gall wasp B. treatae. We did not detect significant differences in the frequency of Wolbachia infection among populations of gall wasps attacking different host plants or among the sexual and asexual generation. Rather the distribution of the three common Wolbachia strains – with wTre1 present exclusively in the western clade and wTre2 and wTre3 (that with the exception of a single individual) being exclusively present in gall wasps from the eastern clade – highlights the role of geography in the Wolbachia infection status of this gall wasp species. The occurrence of different Wolbachia strains within B. treatae populations and their strict association with mitochondrial haplotypes of the host suggests a potential role of the endosymbiont in reproductive isolation in B. treatae.
Abbreviations
- CI:
-
Cytoplasmic incompatibility
- COI:
-
Cytochrome oxidase I
- MLST:
-
Multilocus sequence typing
- mtDNA:
-
Mitochondrial DNA
- Qf :
-
Quercus fusiformis
- Qg :
-
Quercus geminata
- Qv :
-
Quercus virginiana
- SE:
-
Standard error
- wsp :
-
Wolbachia surface protein
- wTre:
-
Wolbachia from B. treatae
References
Makepeace BL, Gill AC. Wolbachia. Rickettsiales Springer International Publishing; 2016. pp. 465–512.
Engelstädter J, Hurst GDD. The ecology and evolution of microbes that manipulate host reproduction. Annu Rev Ecol Evol S. 2009;40:127–49.
Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:2753–63.
Hosokawa T, Koga R, Kikuchi Y, Meng X-Y, Fukatsu T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci U S A. 2010;107:769–74.
Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Micro. 2008;6:741–51.
Bordenstein SR. Symbiosis and the origin of species. In: Bourtzis K, Thomas AM, editors. Insect Symbiosis; 2003. p. 283–304.
Werren JH. Wolbachia and Speciation. In: Berlocher SH, Howerd D, editors. Endless forms. Species and speciation; 1998. p. 245–60.
Hoffmann AA, Turelli M. Cytoplasmatic incompatibility in insects. In: O'Neill SL, Hoffmann AA, Werren JH, editors. Influential passengers: Inherited microorgansisms and Arthropod Reproduction; 1997. p. 42–80.
Turelli M, Hoffmann AA. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature. 1991;353:440–2.
Kriesner P, Hoffmann AA, Lee SF, Turelli M, Weeks AR. Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog. 2013;9:e1003607.
Schuler H, Köppler K, Daxböck-Horvath S, Rasool B, Krumböck S, Schwarz D, et al. The hitchhiker's guide to Europe: the infection dynamics of an ongoing Wolbachia invasion and mitochondrial selective sweep in Rhagoletis cerasi. Mol Ecol. 2016;25:1595–609.
Schuler H, Bertheau C, Egan SP, Feder JL, Riegler M, Schlick-Steiner BC, et al. Evidence for a recent horizontal transmission and spatial spread of Wolbachia from endemic Rhagoletis cerasi (Diptera: Tephritidae) to invasive Rhagoletis cingulata in Europe. Mol Ecol. 2013;22:4101–11.
Turelli M, Hoffmann AA, McKechnie SW. Dynamics of cytoplasmic incompatibility and mtDNA variation in natural Drosophila simulans populations. Genetics. 1992;132:713–23.
Hurst GDD, Jiggins FM. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. P R Soc B. 2005;272:1525–34.
Jiggins FM. Male-killing Wolbachia and mitochondrial DNA: selective sweeps, hybrid introgression and parasite population dynamics. Genetics. 2003;164:5–12.
Raychoudhury R, Baldo L, Oliveira DCSG, Werren JH. Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution. 2009;63:165–83.
Brucker RM, Bordenstein SR. Speciation by symbiosis. Trends Ecol Evol. 2012;27:443–51.
Schuler H, Hood GR, Egan SP, Feder JL. Modes and mechanisms of speciation. In: Meyers RA, editor. Reviews in cell biology and molecular medicine. 3rd ed. Ltd: John Wiley & Sons; 2016. p. 60–93.
Bordenstein SR, O'Hara F, Werren JH. Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature. 2001;409:707–10.
Jaenike J, Dyer KA, Cornish C, Minhas MS. Asymmetrical reinforcement and Wolbachia infection in Drosophila. PLoS Biol. 2006;4:1852–62.
Miller WJ, Ehrman L, Schneider D. Infectious speciation revisited: impact of symbiont-depletion on female fitness and mating behavior of Drosophila paulistorum. PLoS Pathog. 2010;6:e1001214.
Egan SP, Hood GR, Feder JL, Ott JR. Divergent host-plant use promotes reproductive isolation among cynipid gall wasp populations. Biol Lett. 2012;8:605–8.
Egan SP, Hood GR, Ott JR. Testing the role of habitat isolation among ecologically divergent gall wasp populations. International Journal of Ecology. 2012:1–8.
Egan SP, Hood GR, Devela G, Ott JR. Parallel patterns of morphological and behavioral variation among host-associated populations of two gall wasp species. PLoS One. 2013;8:e54690.
Stone GN, Schönrogge K, Atkinson RJ, Bellido D, Pujade-Villar J. The population biology of oak gall wasps (Hymoenoptera: Cynipidae). Annu Rev Entomol. 2002;47:633–68.
Plantard O, Rasplus JY, Mondor G, Le Clainche I, Solignac M. Wolbachia-induced thelytoky in the rose gallwasp Diplolepis spinosissimae (Giraud) (Hymenoptera : Cynipidae), and its consequences on the genetic structure of its host. 1998;265:1075–1080.
Plantard O, Rasplus JY, Mondor G, Le Clainche I, Solignac M. Distribution and phylogeny of Wolbachia inducing thelytoky in Rhoditini and “Aylacini” (Hymenoptera : Cynipidae). Insect Mol Biol. 1999;8:185–91.
Wachi N, Abe Y, Inomata N, Szmidt AE, Tachida H. Speciation history of three closely related oak gall wasps, Andricus mukaigawae, A. kashiwaphilus, and A. pseudoflos (Hymenoptera: Cynipidae) inferred from nuclear and mitochondrial DNA sequences. Mol Ecol. 2012;21:4681–94.
Abe Y, Miura K. Doses Wolbachia induce unisexuality in oak gall wasps? (Hymenoptera: Cynipidae). Ann Entomol Soc Am. 2002;95:583–6.
Rokas A, Atkinson RJ, Nieves-Aldrey JL, West SA, Stone GN. The incidence and diversity of Wolbachia in gallwasps (Hymenoptera; Cynipidae) on oak. Mol Ecol. 2002;11:1815–29.
Hood GR, Ott JR. Developmental plasticity and reduced susceptibility to natural enemies following host plant defoliation in a specialized herbivore. Oecologia. 2010;162:673–83.
Forbes AA, Hall MC, Lund J, Hood GR, Izen R, Egan SP, Ott JR. Parasitoids, Hyperparasitoids, and inquilines associated with the sexual and asexual generations of the gall former, Belonocnema treatae (Hymenoptera: Cynipidae). Ann Entomol Soc Am. 2016;109:49–63.
Ronquist F. Phylogeny, classification and evolution of the Cynipoidea. Zool Scr. 1999;28:139–64.
Abe Y, Melika G, Stone GN. The diversity and phylogeography of cynipid gallwasps (Hymenoptera: Cynipidae) of the oriental and eastern Palearctic regions, and their associated communities. Orient Insects. 2007;41:169–212.
Liljeblad J, Nieves-Aldrey J-L, Neser S, Melika G. Adding another piece to the cynipoid puzzle: the description of a new tribe, genus and species of gall wasp (Hymenoptera: Cynipidae) endemic to the Republic of South Africa. Zootaxa. 2011;2806:35–52.
Lund JN, Ott JR, Lyon RJ. Heterogony in Belenocnema treatae Mayr (Hymenoptera: Cynipidae). Proc Entomol Soc Wash. 1998;100:755–63.
Crozier RH. Hymenoptera. In: John B, editor. Animal cytogenetics, volume 3 Insecta 7. Insecta; 1975;1–95.
Cavender-Bares J, Pahlich A. Molecular, morphological, and ecological niche differentiation of sympatric sister oak species, Quercus virginiana and Q. geminata (Fagaceae). Am J Bot. 2009;96:1690–702.
Cavender-Bares J, González-Rodríguez A, Eaton DAR, Hipp AAL, Beulke A, Manos PS. Phylogeny and biogeography of the American live oaks (Quercus subsection Virentes): a genomic and population genetics approach. Mol Ecol. 2015;24:3668–87.
Braig HR, Zhou W, Dobson SL, O'Neill SL. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J Bacteriol. 1998;180:2373–8.
Arthofer W, Riegler M, Schneider D, Krammer M, Miller WJ, Stauffer C. Hidden Wolbachia diversity in field populations of the European cherry fruit fly, Rhagoletis cerasi (Diptera, Tephritidae). Mol Ecol. 2009;18:3816–30.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotechnol. 1994;3:294–9.
Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Ronquist F, Nieves-Aldrey J-L, Buffington ML, Liu Z, Liljeblad J, Nylander JAA. Phylogeny, evolution and classification of gall wasps: the plot thickens. PLoS One. 2015;10:e0123301.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42.
Huelsenbeck JP, Larget B, Alfaro ME. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol Biol Evol. 2004;21:1123–33.
Lo N, Casiraghi M, Salati E, Bazzocchi C, Bandi C. How many Wolbachia Supergroups exist? Mol Biol Evol. 2002;19:341–6.
R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/.
Xiao J-H, Wang N-X, Murphy RW, Cook J, Jia L-Y, Huang D-W. Wolbachia infection and dramatic intraspecific mitochondrial DNA divergence in a fig wasp. Evolution. 2012;66:1907–16.
Jäckel R, Mora D, Dobler S. Evidence for selective sweeps by Wolbachia infections: phylogeny of Altica leaf beetles and their reproductive parasites. Mol Ecol. 2013;22:4241–55.
O'Neill SL, Giordano R, Colbert A, Karr T, Robertson H. 16S ribosomal-RNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci U S A. 1992;89:2699–702.
Baldo L, Ayoub NA, Hayashi CY, Russell JA, Stahlhut JK, Werren JH. Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol Ecol. 2008;17:557–69.
Gerth M, Röthe J, Bleidorn C. Tracing horizontal Wolbachia movements among bees (Anthophila): a combined approach using MLST data and host phylogeny. Mol Ecol. 2013;22:6149–62.
Egan S, Ott JR. Host plant quality and local adaptation determine the distribution of a gall-forming herbivore. Ecology. 2007;88:2869–79.
Rokas A, Atkinson RJ, Brown GS, West SA, Stone GN. Understanding patterns of genetic diversity in the oak gallwasp Biorhiza pallida: demographic history or a Wolbachia selective sweep? Heredity. 2001;87:294–304.
Yang XH, Zhu DH, Liu Z, Zhao L, Su CY. High levels of multiple infections, recombination and horizontal transmission of Wolbachia in the Andricus mukaigawae (Hymenoptera; Cynipidae) communities. PLoS One. 2013;8:e78970.
Kikuchi Y, Fukatsu T. Diversity of Wolbachia endosymbionts in heteropteran bugs. Appl Environ Microbiol. 2003;69:6082–90.
Ijichi N, Kondo N, Matsumoto R, Shimada M, Ishikawa H, Fukatsu T. Internal spatiotemporal population dynamics of infection with three Wolbachia strains in the adzuki bean beetle, Callosobruchus chinensis (Coleoptera : Bruchidae). Appl Environ Microbiol. 2002;68:4074–80.
Keeling MJ, Jiggins FM, Read JM. The invasion and coexistence of competing Wolbachia strains. Heredity. 2003;91:382–8.
Heath BD, Butcher R, Whitfield W, Hubbard S. Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr Biol. 1999;9(6):313.
Schuler H, Kern P, Arthofer W, Vogt H, Fischer M, Stauffer C, Riegler M. Wolbachia in parasitoids attacking native european and introduced eastern cherry fruit flies in Europe. Environ Entomol. 2016;45:1424–31.
Noda H, Miyoshi T, Zhang Q, Watanabe K, Deng K, Hoshizaki S. Wolbachia infection shared among planthoppers (Homoptera : Delphacidae) and their endoparasite (Strepsiptera : Elenchidae): a probable case of interspecies transmission. Mol Ecol. 2001;10:2101–6.
Schilthuizen M, Stouthamer R. Distribution of Wolbachia among the guild associated with the parthenogenetic gall wasp Diplolepis rosae. Heredity. 1998;81:270–4.
West SA, Cook JM, Werren JH, Godfray H. Wolbachia in two insect host-parasitoid communities. Mol Ecol. 1998;7:1457–65.
Price PW, Abrahamson WG, Hunter MD, Melika G. Using gall wasps on oaks to test broad ecological concepts. Conserv Biol. 2004;18:1405–16.
Melika G, Abrahamson WG. Review of the world genera of oak cynipid wasps. Melinka G, Thuroczy C, editors. Parasitic wasps: Evolution, systematics, biodiversity and. Biol Control. 2002:150–90.
Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–110.
Riegler M, Sidhu M, Miller WJ, O'Neill SL. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr Biol. 2005;15:1428–33.
Kaur R, Siozios S, Miller WJ, Rota-Stabelli O. Insertion sequence polymorphism and genomic rearrangements uncover hidden Wolbachia diversity in Drosophila suzukii and D. subpulchrella. Sci Rep. 2017;7:14815.
Telschow A, Hammerstein P, Werren JH. The effect of Wolbachia on genetic divergence between populations: models with two-way migration. Am Nat. 2002;160:54–66.
Telschow A, Yamamura N, Werren JH. Bidirectional cytoplasmic incompatibility and the stable coexistence of two Wolbachia strains in parapatric host populations. J Theor Biol. 2005;235:265–74.
Atyame CM, Labbe P, Rousset F, Beji M, Makoundou P, Duron O, et al. Stable coexistence of incompatible Wolbachia along a narrow contact zone in mosquito field populations. Mol Ecol. 2015;24:508–21.
Acknowledgements
We thank C. Nice and K. Bell for help with DNA extractions, the EEB discussion group at Texas State, C. Stauffer, T. Micklem and two anonymous reviewers for helpful suggestions.
Funding
This research was supported by the Austrian Science Fund project J-3527-B22 (HS); Faculty Research Enhancement Grants in 2013 and 2015 from Texas State University (JRO); Howard McCarley Student Research Awards from the Southwestern Association of Naturalists (ALD, RWB); Freeman Center Fellows Awards from Texas State University (ALD, RWB); a Theodore Roosevelt Memorial Grant from the American Museum of Natural History (RWB); and the Rice Academy of Fellows at Rice University (GRH). The funders had no role in study design, data collection and analysis, and preparation of the manuscript.
Availability of data and materials
Representative Wolbachia sequences were deposited in GenBank with the accession numbers MG252471–252474 and mitochondrial sequences for all B. treatae haplotypes were deposited in GenBank with the accession numbers MG252379–252470. The raw data generated is available at Dryad: doi:https://doi.org/10.5061/dryad.6k94r.
Author information
Authors and Affiliations
Contributions
HS, SPE, GRH and JRO conceived and designed the studies. HS, GRH, RWB and ALD collected and analyzed the data. HS, SPE, GRH and JRO wrote the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
We have research agreements with Archbold Biological Station in Lake Placid, Florida, Oceola County Parks near Orlando, FL, and the South Florida Water Management District, which manages land surrounding Florida’s Kissimmee River. All other sampling was performed along public roads, where insects were collected and preserved for scientific research purposes only and no permit is required by US law under the authority of 7CFR 330.200.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Infection frequencies of Wolbachia strains in B. treatae populations by host plant, generation and sex. Host = host plant of B. treatae, Gen = generation examined, Sex = female (f), male (m), n = sample size and Wolbachia infection. (XLSX 23 kb)
Additional file 2:
Table S2. Maternal transmission rate of Wolbachia. Infection = infection status of the female, offspring = number of offspring tested for Wolbachia, uninfected = number of uninfected individuals, infected = number of infected individuals, transmission rate = transmission rate of Wolbachia from infected mothers to their offspring. (XLSX 42 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Schuler, H., Egan, S.P., Hood, G.R. et al. Diversity and distribution of Wolbachia in relation to geography, host plant affiliation and life cycle of a heterogonic gall wasp. BMC Evol Biol 18, 37 (2018). https://doi.org/10.1186/s12862-018-1151-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-018-1151-z