Abstract
Background
In the poultry and livestock industries, precise genetic information is crucial for improving economic traits. Thus, functional genomic studies help to generate faster, healthier, and more efficient animal production. Chicken myoblast cells, which are required for muscle development and regeneration, are particularly important because chicken growth is closely related to muscle mass.
Results
In this study, we induced expression of microRNA-146b-5p mediated by the piggyBac transposon system in primary chicken myoblast (pCM) cells. Subsequently, we analyzed and compared the proliferation and differentiation capacity and also examined the expression of related genes in regular pCM (rpCM) cells and pCM cells overexpressing miRNA-146b-5p (pCM-146b OE cells). pCM-146b OE cells showed increased proliferation and upregulated gene expression related to cell proliferation. In addition, next-generation sequencing analyses were performed to compare global gene expression patterns between rpCM cells and pCM-146b OE cells. We found that the higher proliferation in pCM-146b OE cells was the result of upregulation of gene sets related to the cell cycle. Moreover, miRNA-146b-5p overexpression had inhibitory effects on myotube differentiation in pCM cells.
Conclusions
Collectively these results demonstrate that miR-146b-5p is closely related to the proliferation and differentiation of chicken myogenic cells as a modulator of post-transcription.
Similar content being viewed by others
Background
Since the genome sequences of avian species have become available, research has aimed to increase muscle mass, enhance muscle regeneration, and reduce fatty acid accumulation to improve growth. Understanding the genes or genetic markers involved in biological functions and regulatory pathways can help to improve economically important traits in the poultry industry [1,2,3]. Thus, functional genomic studies are powerful and effective for investigating the modulatory mechanisms between cell proliferation and differentiation, in particular in skeletal muscles [4,5,6,7]. Our study was conducted in chicken myoblasts derived from embryonic tissue. Myoblasts are derived from satellite cells, which are a precursor to myogenesis [8]. In the quiescent satellite cell stage, PAX7, a critical marker of undifferentiated myoblasts, is highly expressed. After activation, myoblasts start to proliferate, decrease expression of PAX7, and increase expression of MYOD, a myogenic regulatory factor (MRF). Then they enter the stage of terminal differentiation. In this stage, expression of MYOD decreases, and expression of markers of terminal differentiation such as Myogenin and Desmin increases. Eventually myoblast cells form new myotubes, which in turn form new myofibers [9]. Therefore, myoblasts are closely related to muscle growth, which is an economically important trait of domestic animals.
MicroRNA (miRNA) is a small non-coding RNA molecule that can regulate targeted gene expression by specific mRNA degradation and translational inhibition [10,11,12]. There are numerous reports of miRNAs controlling developmental and cellular processes such as cell proliferation, differentiation, and tissue specification [13, 14]. In addition, some miRNAs, such as miRNA-1 and miRNA-206, control myogenesis in mammals [15, 16]. miRNA-146b (miR-146b) is well conserved in most vertebrates and has many biological functions in innate immunity, inflammation, and cell senescence [17,18,19]. Dicing the pre-miR-146b stem-loop creates two different miRNA species: miR-146b-5p as a major form and miR-146b-3p as a minor form [17,18,19]. miR-146b-5p is a key regulator of muscle regeneration and myoblast differentiation in mice [20]. Furthermore, miR-146b-3p controls myoblast proliferation and differentiation in chicken [21].
However, additional research is required because it is not clear how miR-146b-5p affects myogenic differentiation, and there are no reports addressing its effects on chicken myogenesis. Thus, in this study, we designed and constructed a miRNA expression vector system to overexpress miR-146b-5p in pCM cells using the piggyBac transposon system, which previous studies have demonstrated to be an efficient transgene delivery system [22, 23]. miR-146b-5p increased proliferation and decreased differentiation by regulating genes related to the cell cycle in chicken myoblasts.
Results
miR-146b-5p overexpression in pCM cells
Based on our previous study using a miRNA expression system [24], we designed and constructed a piggyBac transposon-mediated miR-146b-5p overexpression vector (piggyBac CMV-GFP-miRNA-146b-5p; Fig. 1a). Two copies of miR-146b-5p were simultaneously transcribed with the GFP transgene under the CMV promoter (Fig. 1a). This miRNA expression cassette system was used to overexpress the targeted miRNA and also to visualize GFP in the transfected cells. The rpCM and pCM-146b OE cells showed no differences in terms of morphological features (Fig. 1b). Quantitative RT-PCR (qRT-PCR) was performed to determine the overexpression of miR-146b-5p in pCM-146b OE cells. Expression of miR-146b-5p was significantly upregulated in pCM-146b OE cells compared to rpCM cells (Fig. 1c).
Design of the chicken miRNA-146b-5p expression vector and characterization of miR-146b-5p overexpression in primary chicken myoblast (pCM) cells. a The expression vector of piggyBac CMV-GFP-mir146b-5p. The cytomegalovirus and EF1 promoter controlled the expression of GFP-miR146b-5p and the puromycin resistance gene, respectively. b Morphology of regular pCM (rpCM) and pCM cells overexpressing miR-146b-5p (pCM-146b OE cells; scale bar = 100 μm). c mRNA expression profiles of miR-146b-5p were compared between regular pCM and pCM-146b OE cells by qRT-PCR (**p < 0.01)
Characterization of pCM-146b OE cells in an undifferentiated state
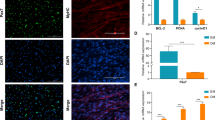
To examine gene expression patterns of the myogenic markers and targets of miR-146b-5p in the undifferentiated state, we compared results of qRT-PCR and Western blotting between rpCM and pCM-146b OE cells. Based on miRBase (http://www.mirbase.org), we selected predicted targets of miR-146b-5p and analyzed their expression patterns in pCM-146b OE cells. All predicted target transcripts (Smad4, Numb, Asck3, Rrm2b, and Sgcb) were significantly downregulated in pCM-146b OE cells compared to rpCM cells (Fig. 2a). Expression of Pax7, a critical marker of undifferentiated myoblasts, was downregulated, whereas expression of the MRF MyoD was upregulated (Fig. 2b). Western blotting confirmed the downregulated and upregulated expression of PAX7 and MYOD, respectively (Fig. 2c). These results indicate that miR-146b-5p may be involved in transcriptional regulation of myogenic genes in pCM cells.
Gene expression analyses of undifferentiated primary chicken myoblast (pCM) cells by quantitative RT-PCR (qRT-PCR) and Western blotting. a mRNA expression profiles of target transcripts of miR-146b-5p (Smad4, Numb, Asck3, Rrm2b, and Sgcb) were compared between regular pCM (rpCM) and pCM cells overexpressing miRNA-146b-5p (pCM-146b OE cells) by qRT-PCR (n = 3; **p < 0.01, ***p < 0.001). b mRNA expression profiles of Pax7 and MyoD were compared between rpCM and pCM-146b OE cells by qRT-PCR (n = 3; **p < 0.01, ***p < 0.001). c Protein expression profiles of PAX7 and MYOD were compared between undifferentiated regular pCM and pCM-146b OE cells. The graph represents a density comparison of Western blotting results (n = 3; *p < 0.05)
Overexpression of miR-146b improves the proliferation of pCM cells
It is intriguing that pCM-146b OE cells had a higher proliferative growth rate. To exclude the influence of GFP, which was inserted into the miR-146b-5p overexpression vectors, we compared proliferation in pCM-GFP cells and pCM-146b OE cells. pCM-146b OE cells had a higher growth rate than pCM-GFP cells after 3 days of in vitro culture (Fig. 3a). In addition, cell cycle analyses with BrdU showed that subsets of pCM-146b OE cells in S phase were significantly increased compared to rpCM cells (rpCM cells: 22.6 ± 0.9% vs. pCM-146b OE cells: 47.8 ± 6.0%, n = 3; Fig. 3a). Furthermore, we analyzed genes related to cell proliferation (Ccnd3, Irf2, Wnt5a, and Pdgfrb) by qRT-PCR. All transcripts, which are positive regulators of proliferation, were upregulated in pCM-146b OE cells (Fig. 3b). These results suggest that miR-146b-5p has effects on the proliferation of skeletal muscle in chicken myoblast cells.
Proliferation and gene expression analyses during primary chicken myoblast (pCM) cell growth. a Cell proliferation was compared between GFP-expressing pCM (pCM-GFP cells) and pCM cells overexpressing miRNA-146b-5p (pCM-146b OE cells) over 5 days (n = 3; **p < 0.01, ***p < 0.001). The right panels show flow cytometry analyses with BrdU for cell cycles, indicating that subsets of pCM-146b OE cells in S phase were significantly increased compared to rpCM cells (rpCM cells: 22.6 ± 0.9% vs. pCM-146b OE cells: 47.8 ± 6.0%, n = 3). b mRNA expression profiles of Ccnd3, Irf2, Wnt5a, and Pdgfrb were compared between regular pCM and pCM-146b OE cells by qRT-PCR (n = 3; **p < 0.01, ***p < 0.001)
Global gene expression analyses by RNA sequencing in pCM-146b OE cells
Next we conducted mRNA sequencing analyses to compare global gene expression patterns between rpCM and pCM-146b OE cells. From mRNA sequencing data, we sorted out a total of 647 DEGs, of which 291 were downregulated and 356 were upregulated (Fig. 4a). Using fold change cutoffs of ≥1.5 (p < 0.001) and ≤ 0.6 (p < 0.001) for upregulated and downregulated gene sets, respectively, we identified four upregulated and two downregulated gene sets in the gene set enrichment analyses (http://software.broadinstitute.org/gsea/index.jsp; Table 1). Subsequently, heatmap analyses for mRNA sequencing data were conducted to visualize the different expression patterns of the predicted target transcripts and genes related to the cell cycle analyzed in Figs. 2 and 3, respectively (Fig. 4c). The gene sets globally up- and downregulated in pCM and pCM-146b OE cells were also visualized (Fig. 4d). To validate the DEGs from mRNA sequencing analyses, we analyzed the gene expression patterns of six upregulated genes (CCNB2, CDC20, KIF23, KPNA2, PLK1, and TOP2A) related to cell cycle regulation. All six transcripts were highly upregulated in pCM-146b OE cells (Fig. 5a). To understand the functional interactions between the upregulated genes and their neighbor genes, we performed STRING analyses (Fig. 5b). The results indicated that miR-146b-5p affected skeletal muscle proliferation and also influenced the regulatory pathways of cell cycling in pCM cells.
mRNA sequencing analyses of regular primary chicken myoblast (rpCM) cells and pCM cells overexpressing miRNA-146b-5p (pCM-146b OE cells). a Differentially expressed genes (DEGs) between the rpCM cells and pCM-146b OE cells were identified with a p cutoff of 0.001 and a fold change cutoff of 1.5. b Scatter plot analyses of DEGs. The red and green dots indicate the expression of upregulated and downregulated genes in the pCM-146b OE cells, respectively. c Heatmap analyses for mRNA sequencing data visualizing the different expression patterns of the predicted target transcripts and genes related to the cell cycle. d The gene sets globally up- and downregulated between rpCM and pCM-146b OE cells were visualized by heatmap analyses
Validation of mRNA sequencing data and string analyses. a Expression profiles for upregulated genes (Ccnb2, Cdc20, Kif23, Kpna2, Plk1, and Top2a) in primary chicken myoblast (pCM) cells overexpressing miRNA-146b-5p (pCM-146b OE cells) by qRT-PCR (n = 3; ***p < 0.001). b String analyses of upregulated gene sets in the pCM-146b OE cells
Overexpression of miR-146b-5p influences myotube differentiation in pCM cells
We compared and analyzed myotube differentiation capacity between rpCM and pCM-146b OE cells. Overexpression of miR-146b-5p dramatically reduced myotube differentiation and formation during myogenesis in pCM-146b OE cells (Fig. 6). pCM-146b OE cells showed fewer differentiated myotubes and less myotube differentiation compared to rpCM cells (Fig. 6a). The area of differentiated myotubes was significantly decreased in pCM-146b OE cells after 4 days of myogenic induction (Fig. 6b). Western blotting showed similar expression patterns in cells in the undifferentiated stage and after the myogenic differentiation (Fig. 7a). Expression of PAX7 was still downregulated, whereas expression of MYOD was upregulated in pCM-146b OE cells. Furthermore, Desmin, a myogenic marker of terminal differentiation [4, 5], was also downregulated in pCM-146b OE cells. In addition, we investigated expression of ID1, which is closely associated with muscle differentiation by binding E proteins (Fig. 7b). Expression of ID1 was significantly upregulated in pCM-146b OE cells. These results demonstrate that overexpression of miR-146b-5p affects the expression of genes associated with myogenic differentiation and support the phenotypic difference between rpCM and pCM-146b OE cells after myogenic differentiation.
Morphological analyses of differentiated pCM cells overexpressing miR-146b (pCM-146b OE). a Morphological comparison of differentiated myotubes of regular pCM (rpCM) and pCM-146b OE cells after 4 days of differentiation. GFP was constantly expressed after myotube differentiation, which suggests that miR146b-5p was also expressed during and after differentiation. White arrows show the differentiated area (scale bar = 100 μm). b Comparison of the percentage of the differentiated area in rpCM and pCM-146b OE cells (n = 3; *p < 0.05)
Protein expression analyses during differentiation and ID1 expression analyses. a Protein expression profiles of PAX7, MYOD, and Desmin were compared between regular pCM (rpCM) cells and pCM cells overexpressing miR-146b (pCM-146b OE) after 4 days of myotube differentiation. The graph represents a density comparison of Western blotting results (n = 3; *p < 0.05). b Expression profiles of ID1 were compared between the rpCM cells and pCM-146b OE cells (n = 3; ***p < 0.001)
Discussion
In this study, we showed that miR-146b-5p regulates myogenic proliferation and differentiation in pCM cells. A previous report suggested that chicken miR-146b-3p regulates myoblast proliferation and differentiation by negatively controlling the PI3K/AKT1 pathway and MDFIC [21]. In addition, mouse miR-146b-5p promotes myogenic differentiation in muscle cells by regulating computationally predicted target genes [20]. Chicken miR-146b-3p and mouse miR-146b-5p show different seed sequences and target genes, which indicates that the functional activity of miRNAs in the previous reports could be different from that of chicken miRNA-146b-5p in chicken muscle cells. To date, there are no data on the influences of chicken miR-146b-5p on the growth and differentiation of myoblast cells. Therefore, we investigated the effects of miR-146b-5p overexpression during myogenic differentiation in pCM cells. In a previous study, the piggyBac transposon was a stable system that integrated the miRNA expression transgene into the chicken genome [22,23,24]. Thus, we used the piggyBac transposon vector to overexpress miR-146b-5p and to deliver genes into the cells. miR-146 OE pCM cells had similar morphological features to rpCM cells but a faster growth rate.
pCM-146b OE cells showed higher expression of genes related to cell proliferation (Ccnd3, Irf2, Wnt5a, and Pdgfrb). The muscle of adult Ccnd3-null mice shows decreased proliferation and impaired G1/S progression, which suggests that CCND3 may be critical for muscle proliferation and cell cycle progression [25]. IRF2 indirectly improves muscle proliferation and regeneration by binding to the VCAM-1 gene promoter in C2C12 cells [26]. Overexpression of WNT5A significantly increases the proliferation of satellite cells derived from mouse muscle and enhances cell division by promoting the nuclear translocation of activated β-catenin [27]. PDGF-induced effects have been observed in C2C12 skeletal muscle cells, resulting in stimulated skeletal muscle proliferation and inhibited myogenic differentiation [28]. Thus, pCM-146b OE cells show that the higher expression of PDGFRB mediated by miR-146b-5p might enhance cell proliferation and inhibit myogenic differentiation. We conducted RNA sequencing in rpCM and pCM-146b OE cells. Based on the RNA sequencing data, we identified upregulated gene sets that were closely related to the cell cycle (Table 1). The transcriptional levels of gene sets related to the cell cycle that were upregulated in pCM-146b OE cells demonstrate that they were reciprocally associated. miR-146b-5p activates and stimulates the regulatory circuits of the myogenic cell cycle and proliferation in pCM cells.
By contrast, myogenic differentiation is suppressed because myotube formation can be induced generally when the cell cycle is arrested [29]. To analyze the functionality of miR-146b-5p during myogenic differentiation, we examined myotube formation patterns and related gene expression after 4 days of differentiation. Although expression of MyoD was upregulated in the differentiated stage and in the undifferentiated stage, pCM-146b OE cells showed a lower differentiation capacity than rpCM cells after 4 days of differentiation (Fig. 6). Similarly, expression of Desmin, a myotube terminal differentiation marker [4, 5], was downregulated in pCM-146b OE cells. Moreover, expression of ID1 protein, an inhibitor of myogenic differentiation in muscle, was upregulated in pCM-146b OE cells (Fig. 7). ID1 protein competitively suppresses the E protein/MYOD complex because it has a higher affinity for the E-protein than MYOD and therefore inhibits myogenic differentiation [30, 31]. mRNA and protein levels of MYOD expression were upregulated, but myotube differentiation was significantly decreased in pCM-146b OE cells (Figs. 6 and 7). It is possible that the upregulation of ID1 protein controlled by miR-146b-5p blocked the E protein/MYOD complex. Furthermore, according to the integrated miRNA annotation and deep-sequencing database (http://www.mirbase.org), SIRT1, a target of miR-146b-5p, helps FOXO transcription factors to bind their target by deacetylation [32, 33]. FOXO3 transcriptionally represses ID1 by directly binding its promoter [34]. This suggests that miR-146b-5p can indirectly control the expression of ID1 protein by inhibiting SIRT1/FOXO3 and the regulatory pathway of myogenic differentiation by regulating the expression of ID1 protein.
Conclusion
In this study, we generated pCM-146b OE cells and conducted a functional assay during myogenic proliferation and differentiation. Compared to rpCM cells, pCM-146b OE cells demonstrated higher proliferation and lower differentiation. pCM-146b OE cells had higher expression of genes related to cell proliferation and the cell cycle. The increased myogenic proliferation suggests that miR-146b-5p can enhance cell proliferation and inhibit myogenic differentiation by regulating the expression of PDGFRB. Furthermore, pCM-146b OE cells demonstrated higher expression of ID1, which indicates that miR-146b-5p can indirectly control myogenic differentiation by regulating the expression of ID1. These results suggest that miR-146b-5p acts as a key regulator of myogenic proliferation and differentiation in chicken.
Methods
Animal care
The procedures for animal management, reproduction, and manipulation adhered to the standard operating protocols of our laboratory at the University Animal Farm, Pyeongchang campus, Seoul National University, South Korea [24].
Primary chicken myoblast (pCM) cell culture and myotube differentiation assay
According to our previous report [24], pCM cells were isolated from the pectoralis major of 10-day-old male chick embryos and maintained in Medium 199 (Invitrogen, Carlsbad, CA, USA; cat#11150–059) supplemented with 10% fetal bovine serum (FBS; HyClone Laboratories, Logan, UT, USA; cat#SH30084.03), 2% chicken serum (Sigma-Aldrich, St. Louis, MO, USA; cat#C5405), and 1× antibiotic-antimycotic (Invitrogen; cat#15240–062) [24]. These cells were cultured in an incubator at 37 °C in an atmosphere of 5% CO2 and 60–70% relative humidity. To differentiate into myotube at 80% confluency of cells, the cells were washed once in phosphate-buffered saline (PBS), and the differentiation medium containing 0.5% FBS and 1× antibiotic-antimycotic was changed. The differentiation medium was replaced with fresh differentiation medium daily according to our previous report [24]. The myotube-differentiated area was measured and analyzed in each well of regular pCM (rpCM) or pCM cells overexpressing miRNA-146b-5p (pCM-146b OE cells) after 4 days of differentiation. All experiments were performed in triplicate with both rpCM or pCM-146b OE cells.
miR146b-5p overexpression vector construction
To overexpress chicken microRNA-146b-5p (miR-146b-5p), we inserted miR-146b-5p into the piggyBac transposon transgene expression system vector (System Biosciences, Palo Alto, CA, USA; cat#PB513B-1) after Asc I digestion and ligation (piggyBac cytomegalovirus [CMV]-GFP-miRNA-146b-5p). The CMV and elongation factor-1 promoters controlled the expression of GFP-miRNA-146b-5p and the puromycin resistance gene, respectively (Fig. 1a). The miRNA-146b-5p was synthesized as 5′-gct ggt gac gtc ccc tat gga att gag ttc tcc gct gtg aca ctt caa act gag aac tga att cca tag gcg atg tgg tca gca-3′ (Bionics, Seoul, Korea).
Development of the miR-146b-5p overexpressing myoblast
For miR-146b-5p-expressing myoblast cells, we conducted co-transfection of the transgene expression vector, piggyBac CMV-GFP (control), or piggyBac CMV-GFP-miRNA-146b-5p with piggyBac transposase using Lipofectamine 3000 (Invitrogen; cat#L3000015) according to our previous report [22]. Transgene DNA-lipid complex consisting of 7.5 μL Lipofectamine 3000 reagent in 250 μL Opti-MEM (Invitrogen; cat#31985–062) and 10 μL P3000 reagent with 2.5 μg piggyBac transgene vector and piggyBac transposase plasmid in 250 μL Opti-MEM was added to each well according to our previous report [22]. One day after lipofection, 10 μg/mL puromycin was added to develop stably transfected cells with the transgene. GFP-expressing cells were observed with a fluorescent microscope (Carl Zeiss Axio Observer A1, Oberkochen, Germany).
Analysis of gene expression by quantitative RT-PCR
Total RNA was extracted with Trizol reagent (Invitrogen; cat#10296010) and cDNA was synthesized with 2 μg RNA and random primers (Invitrogen; cat#18080–051) under standard conditions. Quantitative RT-PCR (qRT-PCR) for miRNA was conducted with the high-specificity miRNA QPCR Core Reagent Kit (Agilent Technology, Santa Clara, CA, USA; cat#600545). Each 20 μL RT-PCR reaction mix contained 2 μL cDNA, 2.5 μL PCR buffer, 1 μL dNTP mixture (2.5 mM), 1 U Taq DNA polymerase, and 10 pmol forward and reverse primers (Table 2). Quantitative RT-PCR analyses were performed with the iCycler iQ Real-time PCR detection system (Bio-Rad, Hercules, CA, USA) and EvaGreen (Biotium, Fremont, CA, USA; cat#31000). The PCR parameters were as follows: an initial incubation at 94 °C for 5 min, followed by 40 cycles at each condition (Table 2). The reaction was terminated by a final incubation at 72 °C for 10 min, and melting curve profiles were analyzed.
Western blotting assay
After extraction with 1× radioimmunoprecipitation lysis buffer, total protein from each treated cells was separated on a 10% polyacrylamide gel and transferred to a nitrocellulose membrane (Bio-Rad). The primary antibodies used were mouse anti-β-actin (Santa Cruz Biotechnology, Dallas, TX, USA; cat#SC-47778), anti-PAX7 (R&D Systems, Minneapolis, MN, USA; cat#MAB1675), anti-MYOD (Santa Cruz Biotechnology; cat#MAB1675), anti-Desmin (Novus Biologicals, Littleton, CO, USA; cat#NB110–1790). Horseradish peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG (Bio-Rad; cat#170–6516 and 170–6515) were used as secondary antibodies. The blots were treated with ECL substrate solutions and exposed in a ChemiDoc XRS System (Bio-Rad) to detect chemiluminescence according to our previous report [24].
Cell growth analyses
For analysis of the cell growth, we subcultured pCM-GFP or pCM-146b OE cells in 24-well culture plates (2 × 104 cells/well). The total number of cells in each well was counted and analyzed statistically during a 5-day in vitro culture. In addition, the proliferative capacities were compared with a 5-bromo-2′-deoxyuridine (BrdU) flow kit (Becton, Dickinson and Company, Franklin Lakes, NJ, USA; cat#559619). Briefly, flow cytometry analyses of cell cycles were compared between rpCM and pCM-146b OE cells after the incorporation of BrdU.
RNA-sequencing and data analyses
To analyze RNA-sequence data in rpCM cells or pCM-146b OE cells, we generated an RNA-sequencing library for each sample and carried out using Illumina HiSeq2500 (Illumina, San Diego, CA). Subsequently, these sequences were aligned and mapped against the chicken reference genome using TopHat for paired-end sequences according to our previous report [7]. Based on searches of the DAVID (http://david.abcc.ncifcrf.gov) and Medline (http://www.ncbi.nlm.nih.gov) databases, we conducted gene classification analysis. We identified differentially expressed genes (DEGs) from rpCM cells and pCM-146b OE cells with a p cutoff of 0.001 and a fold change cutoff of 1.5. Protein–protein association was analyzed with STRING analyses to identify all functional interactions of DEGs (https://string-db.org).
Statistical analyses
We conducted statistical analyses with SAS version 9.4 (SAS Institute, Cary, NC, USA) and the significance of differences was analyzed with a general linear model procedure.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Abbreviations
- miRNA:
-
microRNA
- miR-146b-5p:
-
microRNA-146b-5p
- pCM:
-
Primary chicken myoblast
- rpCM:
-
Regular pCM
- pCM-146b OE:
-
miR-146b-5p overexpressing pCM
- NGS:
-
Next generation sequencing
- MRFs:
-
Myogenic regulatory factors
- GFP:
-
Green fluorescent protein
- CMV:
-
Cytomegalovirus
- qRT-PCR:
-
Quantitative RT-PCR
- Pax7:
-
Paired box protein 7
- MyoD:
-
Myoblast determination protein 1
- SMAD4:
-
SMAD family member 4
- ADCK3:
-
aarF domain containing kinase 3
- RRM2B:
-
Ribonucleotide-diphosphate reductase subunit M2 B
- SGCB:
-
Sarcoglycan Beta
- CCND3:
-
Cyclin D 3
- IRF2:
-
Interferon regulatory factor 2
- PDGFRB:
-
Platelet-derived growth factor receptor beta
- DEGs:
-
Differentially expressed genes
- CCNB2:
-
Cyclin B 2
- CDC20:
-
Cell-division cycle 20
- KIF23:
-
Kinesin-6
- KPNA2:
-
Karyopherin alpha 2
- PLK1:
-
Polo-like kinase 1
- TOP2A:
-
Topoisomerase 2-alpha
- ID1:
-
DNA-binding inhibitor 1
- SIRT1:
-
Sirtuin 1
- FOXO3:
-
Forkhead box O 3
References
International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716.
Claire D'Andre H, Paul W, Shen X, Jia X, Zhang R, Sun L, et al. Identification and characterization of genes that control fat deposition in chickens. J Anim Sci Biotechnol. 2013;4:43.
Rubin CJ, Zody MC, Eriksson J, Meadows JR, Sherwood E, Sherwood E, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–91.
Kim SW, Lee JH, Park BC, Park TS. Myotube differentiation in clustered regularly interspaced short palindromic repeat/Cas9-mediate MyoD knockout quail myoblast cells. Asian-Australas J Anim Sci. 2017;30:1029–36.
Kim SW, Lee JH, Park TS. Functional analysis of SH3 domain containing ring finger 2 during the myogenic differentiation of quail myoblast cells. Asian-Australas J Anim Sci. 2017;30:1183–9.
Lee JH, Kim SW, Park TS. Myostatin gene knockout mediated by Cas9-D10A nickase in chicken DF1 cells without off-target effect. Asian-Australas J Anim Sci. 2017;30:743–8.
Park JW, Lee JH, Kim SW, Han JS, Kang KS, Kim SJ, et al. Muscle differentiation induced up-regulation of calcium-related gene expression in quail myoblasts. Asian-Australas J Anim Sci. 2018;31:1507–15.
Zammit PS, Partridge TA, Yablonka-Reuveni Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J Histchem Cytochem. 2006;54:1177–91.
Olguin HC, Pisconti A. Marking the tempo for myogenesis: Pax7 and the regulation of muscle stem cell fate decisions. J Cell Mol Med. 2012;16:1013–25.
Bartel P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-kappaB. Immunol Rev. 2012;246:205–20.
Izaurralde EGENEREGULATION. Breakers and blockers-miRNAs at work. Science. 2015;349:380–2.
Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–30.
Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–71.
Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–87.
Chen JF, Tao Y, Deng Z, Yan Z, Xiao X, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–79.
Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6.
Bhaumik D, Scott GK, Schokrpur S, Patil CK, Orjalo AV, Rodier F, et al. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY). 2009;(1):402–11.
Perry MM, Williams AE, Tsitsiou E, Larner-Svensson HM, Lindsay MA. Divergent intracellular pathways regulate interleukin-1beta-induced miR-146a and miR-146b expression and chemokine release in human alveolar epithelial cells. FEBS Lett. 2009;583:3349–55.
Khanna N, Ge Y, Chen J. MicroRNA-146b promotes myogenic differentiation and modulates multiple gene targets in muscle cells. PLoS One. 2014. https://doi.org/10.1371/journal.pone.0100657.
Huang W, Guo L, Zhao M, Zhang D, Xu H, Nie Q. The inhibition on MDFIC and PI3K/AKT pathway caused by miR-146b-3p triggers suppression of myoblast proliferation and differentiation and promotion of apoptosis. Cells. 2019. https://doi.org/10.3390/cells8070656.
Park TS, Kim SW, Lee JH. Efficient transgene expression system using a cumate-inducible promoter and Cre-loxP recombination in avian cells. Asian-Australas J Anim Sci. 2017;30:886–92.
Park TS, Park J, Lee JH, Park JW, Park BC. Disruption of G0/G1 switch gene 2 (G0S2) reduced abdominal fat deposition and altered fatty acid composition in chicken. FASEB J. 2019;33:1188–98.
Lee JH, Park JW, Kang KS, Park TS. Forkhead box O3 promotes cell proliferation and inhibits myotube differentiation in chicken myoblast cells. Br Poult Sci. 2019;60:23–30.
De Luca G, Ferretti R, Bruschi M, Mezzaroma E, Caruso M. Cyclin D3 critically regulates the balance between self-renewal and differentiation in skeletal muscle stem cells. Stem Cells. 2013;31:2478–91.
Jesse TL, LaChance R, Iademarco MF, Dean DC. Interferon regulatory factor-2 is a transcriptional activator in muscle where it regulates expression of vascular cell adhesion molecule-1. J Cell Biol. 1998;140:1265–76.
Otto A, Schmidt C, Luke G, Allen S, Valasek P, Muntoni F, et al. Canonical Wnt signaling induces satellite-cell proliferation during adult skeletal muscle regeneration. J Cell Sci. 2008;121:2939–50.
Yablonka-Reuveni Z, Balestreri TM, Bowen-Pope DF. Regulation of proliferation and differentiation of myoblasts derived from adult mouse skeletal muscle by specific isoforms of PDGF. J Cell Biol. 1990;111:1623–9.
Andres V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable event that precede cell fusion upon myogenesis. J Cell Biol. 1996;132:657–66.
Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein id: a negative regulator of helix-loop-helix DNA binding protein. Cell. 1990;61:49–59.
Jen Y, Weintraub H, Benezra R. Overexpression of id protein inhibits the muscle differentiation program: in vivo association of id with E2A protein. Genes Dev. 1992;6:1466–79.
Ahn J, Lee H, Jung CH, Jeon TI, Ha TY. MicroRNA-146b promotes adipogenesis by suppressing the SIRT1-FOXO1 cascade. EMBO Mol Med. 2013;5:1602–12.
Daitoku H, Sakamaki J, Fukamizu A. Regulation of FoxO transcription factors by acetylation and protein-protein interactions. Biochim Biophys Acta. 1813;2011:1954–60.
Birkenkamp KU, Essafi A, van der Vos KE, da Costa M, Hui RC, Holstege F, et al. FOXO3a induces differentiation of Bcr-Abl-transformed cells through transcriptional down-regulation of Id1. J Biol Chem. 2007;282:2211–20.
Acknowledgements
Not applicable.
Authors’ contributions
TSP participated in study design and coordination. JHL participated in the design of the study, carried out the experiments and wrote the first draft of the manuscript. SWK, JSH and SPS carried out and analyzed the experiments. TSP and SIL participated in writing the final version of the manuscript. All authors have read and approved the final manuscript.
Funding
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01334801)” Rural Development Administration, Republic of Korea. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The care and experimental use of chickens were approved by the Institutional Animal Care and Use Committee, Seoul National University (SNU-170417-18-2).
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Western blots in Fig. 2 (C). The original Western blot images of Pax7, MyoD, and β-actin in the undifferentiated stages.
Additional file 2.
Western blots in Fig. 7 (A). The original Western blot images of Pax7, MyoD, Desmin, and β-actin after myotube differentiation.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lee, J.H., Kim, S.W., Han, J.S. et al. Functional analyses of miRNA-146b-5p during myogenic proliferation and differentiation in chicken myoblasts. BMC Mol and Cell Biol 21, 40 (2020). https://doi.org/10.1186/s12860-020-00284-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12860-020-00284-z