Abstract
Background
Unsupervised clustering is a common and exceptionally useful tool for large biological datasets. However, clustering requires upfront algorithm and hyperparameter selection, which can introduce bias into the final clustering labels. It is therefore advisable to obtain a range of clustering results from multiple models and hyperparameters, which can be cumbersome and slow.
Results
We present hypercluster, a python package and SnakeMake pipeline for flexible and parallelized clustering evaluation and selection. Users can efficiently evaluate a huge range of clustering results from multiple models and hyperparameters to identify an optimal model.
Conclusions
Hypercluster improves ease of use, robustness and reproducibility for unsupervised clustering application for high throughput biology. Hypercluster is available on pip and bioconda; installation, documentation and example workflows can be found at: https://github.com/ruggleslab/hypercluster.
Similar content being viewed by others
Background
Unsupervised clustering is ubiquitously used for the interpretation of ‘omics datasets [1,2,3,4,5,6,7]. Clustering is a particularly central challenge in the analysis of single-cell measurement data (e.g. single cell RNA-seq) due to its high dimensionality [8,9,10]. Clustering is also increasingly being used for disease subtype classification and risk stratification [11,12,13,14,15,16,17,18,19]. It is therefore essential that optimal clustering results are easily and robustly obtainable, without user-selected hyperparameters introducing bias and impeding rapid analysis.
Clustering is inherently under-defined [20,21,22]. The definition of “cluster” differs from problem to problem and the desired goal of the analysis [14], and therefore it is not possible to make a single algorithm or metric that can universally identify the “best” clusters [23]. Researchers therefore often compare results from multiple algorithms and hyperparameters [7, 24,25,26,27,28]. Typically, the effect of hyperparameter choice on the quality of clustering results cannot be described with a convex function, meaning that hyperparameters should be chosen through exhaustive grid search [29], a slow and cumbersome process. Software packages for automatic hyperparameter tuning and model selection for regression and classification exist, notably auto-sklearn from AutoML [30], and some groups have made excellent tools for distributing a single clustering calculation for huge datasets [31, 32], but to the best of our knowledge, there is no package for comparing several clustering algorithms and hyperparameters.
Here we present hypercluster, a python package and SnakeMake pipeline for rigorous, reproducible and parallelized clustering calculation and evaluation. This package allows users to compare multiple hyperparameters and algorithms, then easily visualize evaluation metrics for each result [33]. The SnakeMake pipeline allows parallelization, greatly reducing wall-clock time for users [34]. Hypercluster provides researchers with a flexible, parallelized, distributed and user-friendly method for clustering algorithm selection and hyper-parameter tuning.
Implementation
Requirements
The hypercluster package uses scikit-learn [35], python-igraph [36], leidenalg [37] and louvain-igraph [38] to assign cluster labels and uses scikit-learn and custom metrics to compare clustering algorithms and hyperparameters to find optimal clusters for any given input data (Fig. 1). Hypercluster requires python3, pandas [39], numpy [40], scipy [41], matplotlib [42], seaborn [43], scikit-learn [35], python-igraph [36], leidenalg [37], louvain-igraph [38] and SnakeMake [34].
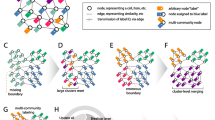
Hypercluster workflow schematic. a Clustering algorithms and their respective hyperparameters are user-specified. Hypercluster then uses those combinations to create exhaustive configurations, and if selected a random subset is chosen. b Snakemake is then used to distribute each clustering calculation into different jobs. c Each set of clustering labels is then evaluated in a separate job by a user-specified list of metrics. d All clustering results and evaluation results are aggregated into tables. Best labels can also be chosen by a user-specified metric.
General workflow and examples
Hypercluster can be run independently of SnakeMake, as a standalone python package. Input and output structure, as well as example workflows on a breast cancer RNA-seq data set [43] and scRNA-seq [45] can be found at https://github.com/ruggleslab/hypercluster/tree/master/examples. Briefly, the workflow starts with instantiating an AutoClusterer (for a single algorithm) or MultiAutoClusterer (for multiple algorithms) object with default or user-defined hyperparameters (Fig. 1a). To run through hyperparameters for a dataset, users simply provide a pandas DataFrame to the “fit'' method on either object (Fig. 1b). Users evaluate the labeling results with a variety of metrics by running the “evaluate” method (Fig. 1c). Clustering labels and evaluations are then aggregated into convenient tables (Fig. 1d), which can be visualized with built in functions (e.g. Additional file 1: Fig. S1, Additional file 2: Fig. S2).
Configuring the SnakeMake pipeline
The SnakeMake pipeline allows users to parallelize clustering calculations on multiple threads on a single computer, multiple compute nodes on a high performance cluster or in a cloud cluster [34]. The pipeline is configured through a config.yml file (Table 1), which contains user-specified input and output directories and files (Table 1, lines 1–3, 5–7) and the hyperparameter search space (Fig. 1a, Table 1, line 18). This file contains predefined defaults for the search space that allow the pipeline to be used “out of the box.” Further, users can specify whether to use exhaustive grid search or random search; if random search is selected, probability weights for each hyperparameter can be chosen (Table 1, line 9). The pipeline then schedules each clustering calculation and evaluation as a separate job (Fig. 1b). Users can specify which evaluation metrics to apply (Fig. 1c, Table 1, line 10) and add keyword arguments to tune several steps in the process (Table 1, lines 4, 8–9, 11–16). Clustering and evaluation results are then aggregated into final tables (Fig. 1d). Users can reference the documentation and examples for more information.
As input, users provide a data table with samples to be clustered as rows and features as columns. Users can then simply run “snakemake -s hypercluster.smk -configfile config.yml” in the command line, with any additional SnakeMake flags appropriate for their system. Applying the same configuration to new files or testing new algorithms on old data simply requires editing the inputs in the config.yml file and rerunning the SnakeMake command.
Extending hypercluster
Currently, hypercluster can perform any clustering algorithm and calculate any evaluation available in scikit-learn [35, 46], as well as non-negative matrix factorization (NMF) [47], Louvain [38] and Leiden [37] clustering. Additional clustering classes and evaluation metric functions can be added by users in the additional_clusterer.py and additional_metrics.py files, respectively, if written to accommodate the same input, outputs and methods (see additional_clusterers.py and additional_metrics.py for examples).
Outputs
For each set of labels, hypercluster generates a file with sample labels and a file containing evaluations of those labels. It also outputs aggregated tables of all labels and evaluations. Hypercluster can also generate several helpful visualizations, including a heatmap showing the evaluation metrics for each set of hyperparameters (Fig. 1c) and a table and heatmap of pairwise comparisons of labeling similarities with a user-specified metric (Additional file 1: Fig. S1). This visualization is particularly useful for finding labels that are robust to differences in hyperparameters. It can also optionally output a table and heatmap showing how often each pair of samples were assigned the same cluster (Additional file 2: Fig. S2). Other useful custom visualizations that are simple for users to create due to the aggregated clustering results are available in our examples (https://github.com/ruggleslab/hypercluster/tree/dev/examples).
Conclusions
Hypercluster allows comprehensive evaluation of multiple hyperparameters and clustering algorithms simultaneously, reducing the allure of biased or arbitrary parameter selection. It also aids computational biologists who are testing and benchmarking new clustering algorithms, evaluation metrics and pre- or post-processing steps [10]. Future iterations of hypercluster could include further cutting-edge clustering techniques, including those designed for larger data sets [31, 32] or account for multiple types of data [48]. Hypercluster streamlines comparative unsupervised clustering, allowing the prioritization of both convenience and rigor.
Availability and requirements
-
Project Name: Hypercluster.
-
Project homepage: https://github.com/ruggleslab/hypercluster/.
-
Operating system: Platform independent.
-
Programming Language: Python.
-
Other requirements: Hypercluster runs with the following versions or higher: python 3.7, pandas 0.24.2, numpy 1.16.4, scipy 1.2.1, matplotlib 3.1.0, seaborn 0.9.0, scikit-learn 0.22.0, hdbscan 0.8.24, snakemake 5.8.2, python-igraph 0.7.1, leidenalg 0.7.0, louvain 0.6.1
-
License: MIT license, open for use by academic and non-academic users.
-
Any restrictions to use by non-academics: Not applicable.
Availability of data and materials
Source code, as well as example vignettes, is available at https://github.com/ruggleslab/hypercluster.
Abbreviations
- NMF:
-
Non-negative matrix factorization
- scRNA-Seq:
-
Single cell RNA-seq
- TCGA:
-
The Cancer Genome Atlas
References
Xu D, Tian Y. A comprehensive survey of clustering algorithms. Ann Data Sci. 2015;2(2):165–93.
Nugent R, Meila M. An overview of clustering applied to molecular biology. Methods Mol Biol. 2010;620:369–404.
Jain AK, Murty MN, Flynn PJ. Data clustering. ACM Comput Surv. 1999;31(3):264–32323.
Xu R, Wunsch DC 2nd. Clustering algorithms in biomedical research: a review. IEEE Rev Biomed Eng. 2010;3:120–54.
Andreopoulos B, An A, Wang X, Schroeder M. A roadmap of clustering algorithms: finding a match for a biomedical application. Brief Bioinform. 2009;10(3):297–314.
Handl J, Knowles J, Kell DB. Computational cluster validation in post-genomic data analysis. Bioinformatics. 2005;21(15):3201–12.
Ronan T, Qi Z, Naegle KM. Avoiding common pitfalls when clustering biological data. Sci Signal. 2016;9(432):re6.
Kiselev VY, Andrews TS, Hemberg M. Publisher Correction: Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet. 2019;20(5):310.
Sun S, Zhu J, Ma Y, Zhou X. Accuracy, robustness and scalability of dimensionality reduction methods for single-cell RNA-seq analysis. Genome Biol. 2019. https://doi.org/10.1186/s13059-019-1898-6.
Liu X, Song W, Wong BY, Zhang T, Yu S, Lin GN, et al. A comparison framework and guideline of clustering methods for mass cytometry data. Genome Biol. 2019;20(1):297.
Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–7.
Ohnstad HO, Borgen E, Falk RS, Lien TG, Aaserud M, Sveli MAT, et al. Prognostic value of PAM50 and risk of recurrence score in patients with early-stage breast cancer with long-term follow-up. Breast Cancer Res. 2017;19(1):120.
Ali HR, Rueda OM, Chin S-F, Curtis C, Dunning MJ, Aparicio SA, et al. Genome-driven integrated classification of breast cancer validated in over 7,500 samples. Genome Biol. 2014;15(8):431.
Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52.
Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–74.
Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164(5):1060–72.
Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173(2):291–304.e6.
Aure MR, Vitelli V, Jernström S, Kumar S, Krohn M, Due EU, et al. Integrative clustering reveals a novel split in the luminal A subtype of breast cancer with impact on outcome. Breast Cancer Res. 2017;19(1):44.
Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52.
Jain AK, Dubes RC. Algorithms clustering data. Old Tappan: Prentice Hall; 1988.
Estivill-Castro V. Why so many clustering algorithms. SIGKDD Explor. 2002;4(1):65–75.
Everitt BS, Landau S, Leese M. Cluster analysis. 4th ed. London: Hodder Arnold; 2001. p. 256.
Von Luxburg U, Williamson RC, Guyon I. Clustering: science or art? Proceedings of ICML Workshop [Internet]. 2012; https://www.jmlr.org/proceedings/papers/v27/luxburg12a/luxburg12a.pdf.
Dhaeseleer P. How does gene expression clustering work? Nat Biotechnol. 2005;23(12):1499–501.
Naegle KM, Welsch RE, Yaffe MB, White FM, Lauffenburger DA. MCAM: multiple clustering analysis methodology for deriving hypotheses and insights from high-throughput proteomic datasets. PLoS Comput Biol. 2011;7(7):e1002119.
Wiwie C, Baumbach J, Röttger R. Comparing the performance of biomedical clustering methods. Nat Methods. 2015;12(11):1033–8.
Rodriguez MZ, Comin CH, Casanova D, Bruno OM, Amancio DR, Costa LF, et al. Clustering algorithms: a comparative approach. PLoS ONE. 2019;14(1):e0210236.
Dubes RC. How many clusters are best? An experiment. Pattern Recognit. 1987;20(6):645–63.
Barber RF, Ha W. Gradient descent with non-convex constraints: local concavity determines convergence. Inf Inference. 2018;7(4):755–806.
Feurer M, Klein A, Eggensperger K, Springenberg J, Blum M, Hutter F. Efficient and robust automated machine learning. In: Cortes C, Lawrence ND, Lee DD, Sugiyama M, Garnett R, editors. Advances in neural information processing systems 28. Red Hook: Curran Associates Inc.; 2015. p. 2962–2970.
Corizzo R, Pio G, Ceci M, Malerba D. DENCAST: distributed density-based clustering for multi-target regression. J Big Data. 2019. https://doi.org/10.1186/s40537-019-0207-2.
Hu X, Liu L, Qiu N, Yang D, Li M. A MapReduce-based improvement algorithm for DBSCAN. J Algorithm Comput Technol. 2018;12(1):53–61.
Van Craenendonck T, Blockeel H. Using internal validity measures to compare clustering algorithms. Benelearn 2015 Poster presentations (online). 2015;1–8.
Köster J, Rahmann S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics. 2012;28(19):2520–2.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–30.
Csardi G, Nepusz T, et al. The igraph software package for complex network research. Int J Complex Syst. 2006;1695(5):1–9.
Traag V. leidenalg [Internet]. Github; [cited 2020 Jan 27]. https://github.com/vtraag/leidenalg
Traag V. louvain-igraph [Internet]. Github; [cited 2020 Jan 27]. https://github.com/vtraag/louvain-igraph
McKinney W, Others. Data structures for statistical computing in python. In: Proceedings of the 9th Python in Science Conference. Austin, TX; 2010. p. 51–6.
van der Walt S, Colbert SC, Varoquaux G. The NumPy array: a structure for efficient numerical computation. Comput Sci Eng. 2011;13(2):22–30.
Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D et al. SciPy 1.0--Fundamental Algorithms for Scientific Computing in Python [Internet]. arXiv [cs.MS]. 2019. https://arxiv.org/abs/1907.10121
Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9(3):90–5.
Waskom M, Botvinnik O, O’Kane D, Hobson P, Lukauskas S, Gemperline DC et al. mwaskom/seaborn: v0.8.1 (September 2017) [Internet]. 2017. https://zenodo.org/record/883859.
Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534(7605):55–62.
Tikhonova AN, Dolgalev I, Hu H, Sivaraj KK, Hoxha E, Cuesta-Domínguez Á, et al. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569(7755):222–8.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. 2.3. Clustering—scikit-learn 0.22 documentation [Internet]. 2011 [cited 2019 Dec 23]. https://scikit-learn.org/stable/modules/clustering.html
Chalise P, Fridley BL. Integrative clustering of multi-level ’omic data based on non-negative matrix factorization algorithm. PLoS ONE. 2017;12(5):e0176278.
Barracchia EP, Pio G, D’Elia D, Ceci M. Prediction of new associations between ncRNAs and diseases exploiting multi-type hierarchical clustering. BMC Bioinform. 2020;21(1):70.
Acknowledgements
We thank the members of Ruggles and Fenyö labs for their helpful discussions and input. We would like to thank MacIntosh Cornwell for his advice with the SnakeMake pipeline. We would also like to thank Joseph Copper Devlin for his help and advice with implementing Louvain and Leiden clustering.
Funding
This work has been supported by the National Cancer Institute (NCI) through CPTAC award U24 CA210972 (PI: David Fenyo) which funded the development of our workflow for the purpose of cancer proteogenomic discovery. Dr. Fenyo oversees the broader CPTAC pancancer projects that highlighted the need for this tool.
Author information
Authors and Affiliations
Contributions
LB and KVR conceived of the ideas and wrote the manuscript. LB developed the method and wrote the code. KVR supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1
. Figure S1: Pairwise label comparisons. Automatically generated heatmap showing pairwise comparison of labeling automatically generated using hypercluster of breast cancer samples. Colors represent adjusted rand index between labels.
Additional file 1
. Figure S2: Pairwise sample comparisons. Automatically generated pairwise comparison of breast cancer samples. Color indicates the number of times two samples were assigned the same cluster.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Blumenberg, L., Ruggles, K.V. Hypercluster: a flexible tool for parallelized unsupervised clustering optimization. BMC Bioinformatics 21, 428 (2020). https://doi.org/10.1186/s12859-020-03774-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12859-020-03774-1